
NEXTFLEX RiboNaut rRNA Depletion Kit (Human / Mouse / Rat)



| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | リボデプレーション |



Product information
Overview
- Designed for human, mouse, and rat total RNA samples, it removes both cytoplasmic and mitochondrial ribosomal RNA.
- Broad transcript detection: Preserves mRNA and non-poly(A) RNA species for total RNA-seq and differential expression studies.
- Flexible input: From 5 ng to 1 µg total RNA.
- Straightforward workflow: Subtractive hybridization with biotinylated probes and magnetic-bead capture.
- Seamless downstream integration: Compatible with essentially any RNA-seq library-prep system, including NEXTFLEX Rapid Directional RNA-Seq 2.0. subsequent libraries can be sequenced on any platform.
- Throughput ready: Works manually or on common liquid handlers, including Sciclone™ and Zephyr™ workstations.
Additional product information
HMR rRNA depletion via subtractive hybridization
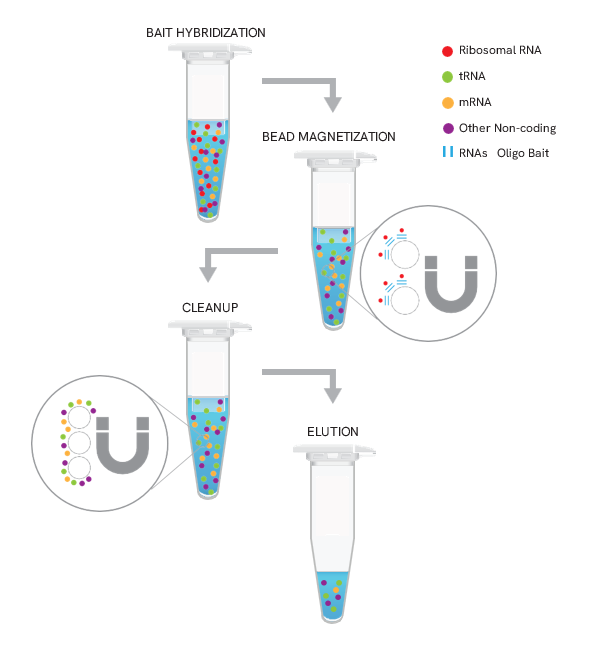
The NEXTFLEX RiboNaut rRNA Depletion Kit (Human, Mouse, Rat) uses a subtractive hybridization workflow to remove ribosomal RNAs, which constitute ~ 80 - 90% of total RNA preparations. A mixture of biotinylated oligonucleotides complementary to cytoplasmic and mitochondrial rRNA in human, mouse, and rat total RNA samples is hybridized to the sample, then captured with streptavidin-coated magnetic beads to pull down rRNA:probe complexes. The supernatant contains rRNA-depleted RNA suitable for total RNA-seq with detection of mRNA and non-poly(A) transcripts. The protocol is optimized for 5 ng to 1 µg total RNA and integrates seamlessly with any RNA-seq library-prep workflow.

Figure 1. Workflow for HMR rRNA depletion with the NEXTFLEX RiboNaut kit. Biotinylated baits hybridize to cytoplasmic and mitochondrial rRNA in human, mouse, and rat. Streptavidin magnetic beads capture rRNA:probe complexes, followed by magnetization, cleanup, and elution to yield rRNA-depleted RNA ready for RNA-seq library prep.
When to use rRNA depletion
For broad transcriptome profiling, rRNA depletion removes 5S, 5.8S, 18S, 28S (cytoplasmic) and 12S, 16S (mitochondrial) rRNA while preserving mRNA and non-poly(A) RNAs, standardizing inputs for total RNA-seq across human, mouse, and rat. Use rRNA depletion when RNA is partially degraded or low, when you need lncRNA and other non-poly(A) species, or when tissues are ribosome rich. Choose poly(A) selection such as NEXTFLEX Poly(A) Beads 2.0 kit when the study targets polyadenylated mRNA from high-quality RNA. For background on how rRNA removal improves total RNA-seq coverage and detection of non-poly(A) transcripts, see our blog from birth to decay: a multi-layer view of the mRNA lifecycle.
Performance metrics after ribodepletion
The NEXTFLEX RiboNaut rRNA Depletion Kit (Human, Mouse, Rat) delivers consistent performance for total RNA-seq after depletion. Example datasets show uniform gene body coverage and minimal residual rRNA contamination across human, mouse, and rat total RNA. Inputs from 5 ng to 1 µg generate rRNA-depleted RNA for stranded RNA-seq library prep.
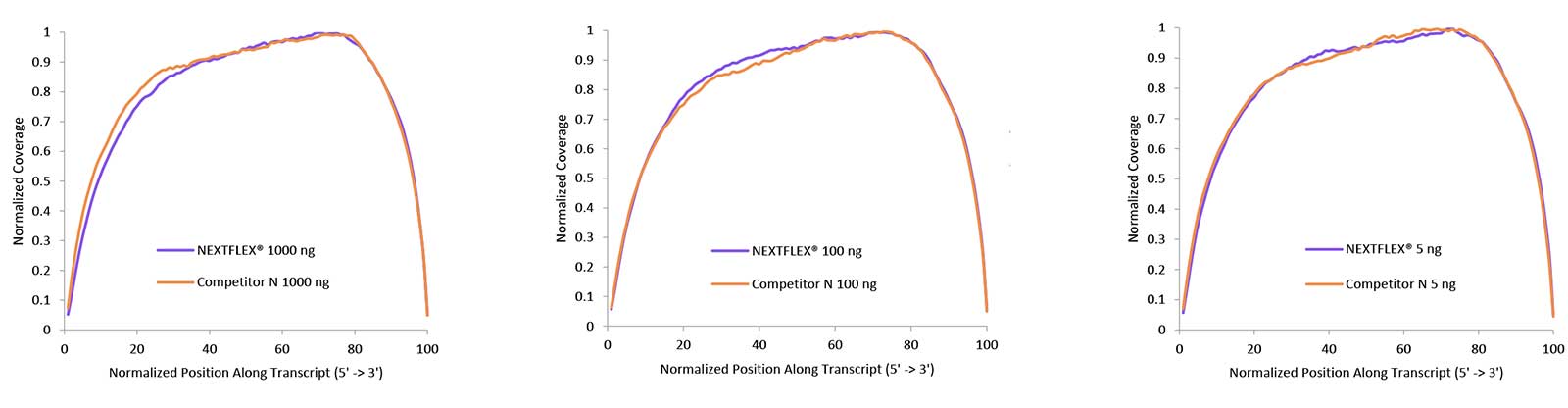
Figure 2. Uniform gene body coverage after HMR rRNA depletion. rRNA-depleted total RNA prepared with the NEXTFLEX RiboNaut rRNA Depletion Kit (Human, Mouse, Rat) shows even coverage along transcripts from 5′ to 3′ at 1000 ng, 100 ng, and 5 ng inputs. Libraries were constructed with NEXTFLEX Rapid Directional RNA-Seq 2.0 and sequenced to a common read depth, yielding overlapping coverage profiles across inputs.
Uniform gene body coverage indicates that depletion and library construction introduce minimal bias, which supports accurate total RNA-seq quantification and isoform analysis. Consistency at low input strengthens performance for scarce or partially degraded HMR samples.
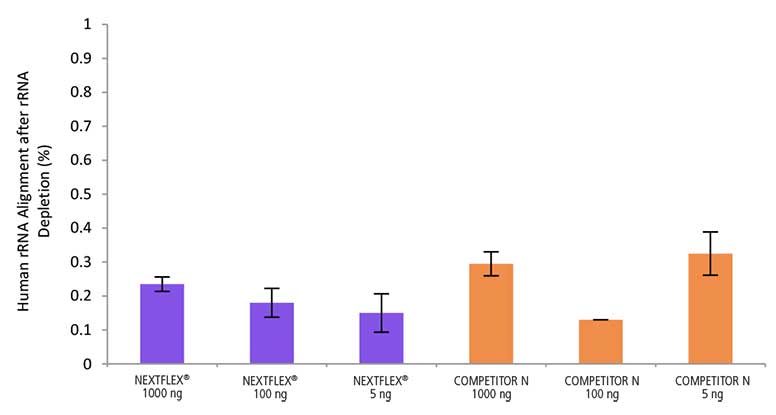
Figure 3. Low residual rRNA after HMR rRNA depletion. Total RNA (Universal Human Reference RNA) was processed with the NEXTFLEX RiboNaut rRNA Depletion Kit (Human, Mouse, Rat). Libraries were prepared using the NEXTFLEX Rapid Directional RNA-Seq 2.0 kit and sequenced on an Illumina® MiSeq® (1×151) platform. Low levels were observed across 5S, 5.8S, 12S, 16S, 18S, and 28S rRNA species, covering both cytoplasmic and mitochondrial rRNA.
Low residual rRNA means more usable reads for genes and non-poly(A) transcripts in total RNA-seq, improving sensitivity and quantification. Reducing both cytoplasmic and mitochondrial rRNA supports consistent performance across HMR samples and input amounts.
Seamless integration with the NEXTFLEX RNA ecosystem for total RNA-seq
The NEXTFLEX RiboNaut rRNA Depletion Kit (Human, Mouse, Rat) plugs into Revvity's broader NEXTFLEX RNA-seq accessory portfolio , After subtractive hybridization and magnetic capture, the resulting rRNA-depleted RNA moves directly into NEXTFLEX Rapid Directional RNA-Seq 2.0 . Indexing can be scaled from pilot runs to 384-plex using UDI or UDI-UMI plates that are color balanced and QC verified to help limit index misassignment in large pools. The end-to-end workflow supports 5 ng to 1 µg inputs and scales from low-input discovery to high-throughput studies on Illumina® and Element® systems.
Automation support
Validated scripts for Revvity Sciclone and Zephyr workstations standardize hybridization, bead capture, and cleanup to reduce hands-on time and variability. Volumes and steps are automation friendly, and the rRNA-depleted RNA transfers directly into automated NEXTFLEX library preparation while maintaining rRNA removal efficiency and strandedness.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Format |
Automation Friendly Volumes
|
| Product Group |
Ribodepletion
|
| Shipping Conditions |
Dual Temperature
|
| Unit Size |
96 rxns
|
Citations
- Laudadio I, Carissimi C, Scafa N, Bastianelli A, Fulci V, Renzini A, et al. (2024). Characterization of patient-derived intestinal organoids for modelling fibrosis in Inflammatory Bowel Disease. Inflammation Research 73(8):1359–1370. DOI: 10.1007/s00011-024-01901-9
- Garcia-del Rio DF, Derhourhi M, Bonnefond A, Leblanc S, Guilloy N, Roucou X, et al. (2024). Deciphering the ghost proteome in ovarian cancer cells by deep proteogenomic characterization. Cell Death & Disease 15(9):712. DOI: 10.1038/s41419-024-07046-1
FAQs
-
What does the NEXTFLEX RiboNaut HMR kit remove?
-
Which species are supported?
-
What RNA input amounts are supported?
-
Does it work with partially degraded RNA or FFPE?
-
Which transcripts are retained after depletion?
-
Which library-prep kits are compatible?
-
Should I choose rRNA depletion or poly(A) selection?
Resources
Are you looking for resources, click on the resource type to explore further.


How can we help you?
We are here to answer your questions.






























