Next‑generation sequencing (NGS) has transformed our understanding of gene expression by giving us genome‑wide, quantitative snapshots of RNA at every checkpoint of its lifecycle. Classical RNA‑seq provides steady-state information about the accumulated, relatively stable population of mature mRNAs present in the sample. In contrast, nascent transcriptomics focuses on detecting newly synthesized RNA molecules still associated with RNA Polymerase II (Pol II) at the site of transcription, while ribosome‑bound RNA footprinting exposes which transcripts actively engage with the translation machinery1–3.
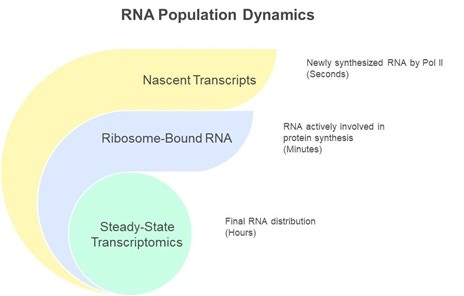
Each assay interrogates a distinct kinetic window, and their biological stories can diverge sharply, even for the same gene (Figure 1). Appreciating those differences is essential when designing or interpreting multi-omic NGS experiments. This blog provides a high-level overview of what each window reveals.

Figure 1. Diagram illustrating the kinetic windows captured by nascent transcriptomics, ribosome profiling and steady-state assays.
Nascent transcriptomics: listening to Pol II breathe
Nascent transcriptomics allows researchers to identify genes that are actively being transcribed at a specific time point, minimizing the influence of post-transcriptional regulation (e.g., RNA processing, stability). For a detailed description of the methods currently used in this area, please refer to our recent blog on this topic. Here we will focus on events that can be uniquely captured when looking at this kinetic window:
- Burst frequency and promoter pausing. Accumulations of Pol II 30–60 nt downstream of the transcription start site quantify how often a gene fires and how long it idles; immediate‑early genes such as FOS display frequent bursts but pause only briefly before elongation resumes3.
- Divergent initiation. Nascent reads expose antisense starts flanking active promoters (e.g. upstream of MYC). These transcripts are rapidly terminated and degraded and therefore invisible in classical RNA‑seq4.
- Enhancer ignition. Short‑lived enhancer RNAs, such as those generated at the β‑globin LCR, rise within minutes of stimulus and track the activation trajectory of their target genes, providing a real‑time read‑out of enhancer‑promoter communication5,6.
- Early termination & attenuation. Polymerase pile‑ups just downstream of cryptic poly(A) motifs (e.g. within NR4A1) flag attenuation checkpoints long before mRNA decay equilibrates, unveiling an additional layer of transcriptional control7.
The RNA obtained in nascent transcriptomics experiments is usually 30-100 nt in length and therefore is processed with special workflows such as the NEXTFLEX™ Small RNA-seq Kit
 NEXTFLEX Small RNA Sequencing Kit V4
8 .
NEXTFLEX Small RNA Sequencing Kit V4
8 .
Ribosome-bound RNA: decoding the cell's investment portfolio
The analysis of RNA fragments bound to ribosomes is known as ribosome profiling or Ribo-Seq2. Ribo-seq provides a snapshot of the pool of RNA that is being actively used to synthesize proteins. It allows researchers to identify translated regions of the genome, quantify translational efficiency, and study the regulation of translation. It is influenced by both transcription and mRNA stability9,10. Please check our recent blog if you are interested in description of the protocols currently used to isolate RNA fragments for Ribo-Seq
 NEXTFLEX Small RNA Sequencing Kit V4
. The events that can be uniquely captured when looking at this kinetic window are the following:
NEXTFLEX Small RNA Sequencing Kit V4
. The events that can be uniquely captured when looking at this kinetic window are the following:
- Translation efficiency. Comparing ribosome footprints to mRNA abundance ranks transcripts by real protein output; for example, during the integrated‑stress response, ATF4 gains a four‑fold translation efficiency boost even as its mRNA level remains flat2.
- Hidden ORFs and micro‑proteins. Footprints CAN uncover upstream ORFs that gate translation (e.g. the two inhibitory uORFs in the ATF4 5′ UTR) and hundreds of sORFs encoding functional micro‑proteins such as the 58‑aa mitochondrial peptide MitoREGULIN that enhances oxidative phosphorylation11.
- Elongation pausing & quality control. Polybasic sequences (e.g., stretches of lysine/arginine) cause ribosome stalling, which leads to Hel2‑mediated ubiquitination of the ribosome and activation of the ribosome associated quality control factors12.
- Isoform specific ORF usage. When alternative splicing changes coding potential, Ribo seq reveals which isoforms reach ribosomes13.
Ribosome protected RNA fragments are usually 28-30 nt in length and therefore can be sequenced downstream with special workflows such as the NEXTFLEX™ Small RNA-seq Kit
 NEXTFLEX Small RNA Sequencing Kit V4
.
NEXTFLEX Small RNA Sequencing Kit V4
.
Steady state bulk: the long game
Classical RNA seq, where the sample is enriched using Poly(A) beads or rRNA depletion remains considered the main method for whole gene expression, splicing, SNV calling and allele specific analyses1. Solutions such as the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 can be used for steady-state analysis of the transcriptome, regardless of input quality. The events that can be uniquely captured when looking at this kinetic window are the following:
- Genome wide quantitative expression. RNA seq can detect > 20,000 genes simultaneously; the GTEx Atlas uses this depth to map tissue specific signatures across 54 human organs14.
- Alternative splicing landscapes. Junction reads uncover exon skipping, such as the switch from anti apoptotic BCL xL to pro apoptotic BCL xS during erythroid maturation15.
- RNA editing maps. Systematic A to I mismatches pinpoint editing hotspots, exemplified by the oncogenic recoding of AZIN116.
- Variant and fusion discovery. Deep coverage enables detection of expressed SNVs and oncogenic fusions17.
Conclusions
No single sequencing assay covers all “transcription”. Nascent data capture initiation and early elongation, ribosome footprints the commitment to protein synthesis, and steady state RNA the integrated outcome of synthesis and decay. Combining at least two layers frequently distinguishes causal from correlative regulation and prevents mis interpretation of fold changes that can arise from buffering mechanisms. As methodological approaches mature, the challenge shifts from generating data to designing questions that exploit each layer's unique temporal window.
References:
- Mortazavi, A., et al. (2008). Mapping and quantifying mammalian transcriptomes by RNA seq. Nat. Methods. 5, 621 628.
- Ingolia, N.T., et al. (2009). Genome wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324, 218 223.
- Core, L. J., et al. (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 322, 1845 1848.
- Schwalb, B., et al. (2016). TT seq maps the human transient transcriptome. Science. 352, 1225 1228.
- Mimoso, C.A., Goldman, S. R. (2023). PRO seq: precise mapping of engaged RNA polymerase II at single nucleotide resolution. Curr. Protoc. 3, e961.
- Bressin, A. et al. (2023). High sensitive nascent transcript sequencing reveals BRD4 specific control of widespread enhancer and target gene transcription. Nat. Commun. 14, 4971.
- Guo, H. et al. (2021). NR4A1 regulates expression of immediate early genes, suppressing replication stress in cancer. Mol. Cell. 81(19): 4041-4058.
- Ting, M.K.Y., et al. (2024). Optimization of ribosome profiling in plants including structural analysis of rRNA fragments. Plant Methods.20: 143.
- Tierney, J.A.S., et al. (2024). RiboSeq.Org: an integrated suite of resources for ribosome profiling data analysis and visualisation. Nucleic Acids Res. 53, D268 D277.
- Tomuro, K., Iwasaki, S. (2025). Advances in ribosome profiling technologies. Biochem. Soc. Trans. 52, BST20253061.
- Averina, O.A., et al. (2023). Mitochondrial peptide Mtln contributes to oxidative metabolism in mice RNA Biochimie. 204:, 136 139.
- Matsuo, Y., et al. (2017). Ubiquitination of stalled ribosome triggers ribosome-associated quality control.. Nat. Comm. 8, 159.
- Reixachs-Solé, M., et al. (2020). Ribosome profiling at isoform level reveals evolutionary conserved impacts of differential splicing on the proteome. . Nat. Comm. 11, 1768.
- GTEx Consortium. (2020). The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 369, 1318 1330.
- Tallack, M.R., et al. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seqGenome Res. 22(12):2385 2398
- Chen L et al. (2023). Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Commun. 14, 1592.
- Heyer, E.E. et al. (2019). Diagnosis of fusion genes using targeted RNA sequencing. Nat. Commun. 10, 1388.