When an Illumina sequencer photographs the flow-cell during an index read, every fluorescent spot becomes a landmark that the software uses to maintain spatial registration across imaging cycles. These landmarks are essential to keep track of the position of thousands of DNA clusters at once. If one imaging channel is suddenly starved of light because every library in the pool shows the same non-fluorescent base at that cycle, the software struggles to align the new frame with the previous one. Registration can drift, phasing models degrade, and the index read quality scores plummet. As a result, affected reads accumulate in the Undetermined bin. Because index reads are used to demultiplex samples, even a small failure in index Q-scores leads to data loss across entire samples. Even worse on instruments with tighter optical tolerances the run can abort. This dependence on per-cycle signal distribution is what Illumina calls color balance, and mastering it is one of the most cost-effective safeguards in next-generation sequencing1.
While color balance also plays a role during insert reading cycles, the impact is typically buffered by the greater base diversity present in the biological sample, where most insert reads contain a mix of A, C, G, and T bases, ensuring enough signal for alignment and phasing models. In contrast, index reads are short (commonly 6–10 bp) and often identical across libraries at the first few positions. This makes them highly vulnerable to imbalanced base composition and channel dropout.
From four channels to one: how the chemistry evolved
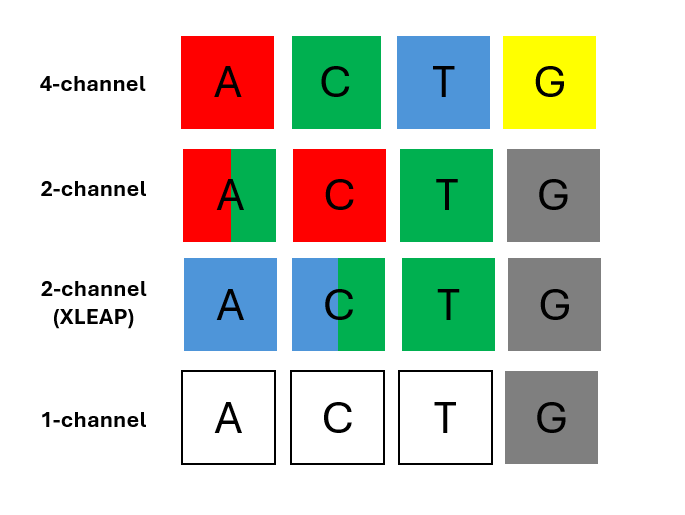
On early Genome Analyzers, Illumina® HiSeq® instruments, each of the four DNA bases was tagged with a unique, base-specific fluorescent dye, emitting light at a different wavelength that was captured in 4 different channels This system, which is still in use in the Illumina® MiSeq® instrument, allows direct detection of each base, making calling robust and tolerant to low diversity libraries. In these instruments color balance was helpful but rarely fatal when ignored.
To increase imaging speed on each cycle, Illumina transitioned to systems with fewer channels. A 4-channel system requires four separate images per cycle, whereas a two-channel system requires only two. This not only helps to reduce run times, critical for high throughput platforms, but also reduces the optical and mechanical complexity of the instrument. Additionally, fewer dyes reduce the cost of regents, supporting more affordable sequencing.
One of the trade-offs of this approach is that reduced-channel count requires usage of combinations or colors for base-calling, increasing software complexity. For example, the two-channel chemistry introduced on the Illumina® MiniSeq®, NextSeq® 500/550 and NovaSeq® 6000 instruments, is based on red and green dyes. In this configuration A is encoded with a combination of red and green, C with red, T with green, and G is unlabeled and produces no signal (“dark” base)
Illumina’s XLEAP® chemistry implemented in the NextSeq® X/X plus system also uses two dyes, blue and green. A is encoded with blue, C blue and green), T remains green, and G is still dark.2
Illumina’s iSeq® 100 goes further, using a single dye imaged twice per cycle, under different excitation and emission conditions. A is identified if fluorescence appears only in the first image, C fluoresces only in the second, T fluoresces in both and G is a dark base, so does not produce signal in either image (Figure 1).

Figure 1. Colors assigned to each base in different instrument types.
Why imbalance causes trouble
Each index cycle begins with a low-magnification pre-scan (often called a survey shot), followed by high-resolution imaging in the active channels. During this process, the base-calling software aligns the new images to the previous cycle by matching the constellation of bright clusters, much like overlaying two photographs of star fields. This alignment is essential to track the position of each DNA cluster across cycles.
But if one or more imaging channels carry no signal, for example, when all libraries have the same base at a certain cycle and that base is dark in the imaging scheme, the constellation of points shrinks or disappears. Alignment becomes approximate, intensities are mis-registered, and the errors snowball. Once the positional uncertainty exceeds a threshold, raw intensities are mis-assigned, Q-scores collapse, and tens of thousands of reads can become undecodable1. This is most critical during Index Read 1 and Index Read 2, especially in the first few cycles, where the signal is used both for cluster registration and color matrix normalization. See Table 1 below for additional information on the sensitivity to imbalance of each detection system.
| Instrument | Detection scheme | Imbalance sensitivity |
|---|---|---|
|
MiSeq® MiSeq® i100 |
4-channels | Even if all sequences begin with the same base, there is still fluorescence, and positional tracking works. Runs usually finish. |
|
MiniSeq® NextSeq® 500/550 NovaSeq® 6000 |
2-channels | Pools in which all i7 starts with GGG across, therefore first three cycles are dark. This creates three black frames, leading to poor registration, no signal for normalization, and ultimately index read failure. |
|
NextSeq® 1000/2000 NovaSeq® X/X Plus |
2-channels (XLEAP) | Also susceptible to G-starting index imbalance, though software improvements in XLEAP increase tolerance slightly. Still, if all samples start with G in i7 or i5, expect frame dropout and index Q-score collapse |
| iSeq® 100 | 1-channel | Dark frames affect both images, so tolerance is lower. If the pool starts with GG, the system sees two completely dark images, loses spatial registration, and index reads are essentially lost. |
Table 1. Sensitivities to color imbalance for all Illumina® instruments currently available
How modern index kits are designed
Commercial index plates are engineered to make color balancing almost automatic. Modern Unique Dual Index (UDI) plates place barcode pairs so that any single column (or row, depending on the vendor) provides a safe mix of fluorescent states across the early cycles, even on two-channel instruments. Illumina’s UD Index plates, for example, enforce recommended pairings and orient the i5 sequences so that users can copy them directly into the sample sheet without breaking balance rules3.
Revvity and other vendors design and test new plates considering both standard two-channel and XLEAP® reagents. The updated XLEAP® color map demands that each cycle contain at least one C or T and one C or A to populate the dual-color channel; plates that fail this test are rearranged before release.2 For very low-plex kits, common in amplicon sequencing, manufacturers sometimes bundle fixed two-, three- or four-plex mixes that have been explicitly vetted for color diversity so users cannot create an unsafe combination by accident. NEXTFLEX™ UDI adapters are carefully curated to maintain color diversity across sequencing cycles, with each set designed and verified for compatibility with the color balance requirements of the different Illumina systems (Table 2). The index sequences are distributed to avoid dark or monochromatic cycles, and the adapter pairs are supplied in a format that minimizes the risk of user-induced imbalance during library prep setup. This design makes these kits especially robust for high-throughput workflows where custom barcode pooling is impractical.
| Detection scheme | Color balance requirement |
|---|---|
| 4-channels | More tolerant. Still avoid mono-base cycles |
| 2-channels | Each cycle: at least one red (A or C) and green (A or T) |
| 2-channels (XLEAP) | Each cycle: at least one blue (A or C) and green (C or T) |
| 1-channel | Each cycle: includes A (Image 1) and C or T (Image 2) |
Table 2. Color balance requirements of the different detection schemes provided by Illumina.
Designing a balanced pool today
Information supplied with commercial adapters should include instructions on which barcodes to use depending on the number of samples you are pooling. Well-designed barcodes allow both low- and high-pooling options without wasting any of the adapters. If this information is not included with the adapters, you can use the following instructions.
For one or two libraries inspect the first three bases of each barcode. On any two- or one-channel instrument the golden rule is simple: never let every library show the same dark or single-color state in the same cycle. Pairing N701 with N702 from a classic Illumina® TruSeq® plate, for instance, guarantees both red and green signal on the first cycle. In the case of NEXTFLEX Unique Dual Index (8 nt index)
 NEXTFLEX Unique Dual Index Barcodes (8NT index, 1-96)
, combinations of barcodes for low level multiplexing are provided directly in the manual.
NEXTFLEX Unique Dual Index Barcodes (8NT index, 1-96)
, combinations of barcodes for low level multiplexing are provided directly in the manual.
When pooling four to twenty-four libraries, aim for rough equality among fluorescent states in the first three cycles. A practical shortcut is to choose an entire column from a 96-well UDI plate and, if you need more samples, add a column from the opposite side; plate layouts are pre-balanced that way. Before the run, paste the chosen indices into Illumina Experiment Manager or run bcl-convert using the “--validate-balance” option to check per-cycle coverage.1
For high-plex pools usually risk if imbalance is very low but may be needed to count bases explicitly. On two-channel systems, make sure both channels carry signals in every cycle, in practice that means at least one C or T in every index cycle. A small Python script (or the validation mode of bcl-convert) can flag weak cycles. Swap in alternative indices that supply the missing base or split the pool across two lanes.2
Some assays, especially highly conserved amplicon panels, cannot meet the rule. For very low-diversity runs on iSeq® instruments, spike-in ≥ 5% PhiX to restore the missing channel signal.4 Confirm the i5 orientation: it is read forward on MiSeq®, MiSeq® i100, and the MiniSeq® (with Rapid reagents), instruments but in reverse-complement on patterned-flow-cell instruments such as iSeq®, MiniSeq® (with Standard reagents), NextSeq®, NovaSeq® 6000 and X series instruments.5
Confirm the i5 orientation: it is read forward on MiSeq®, MiSeq® i100, and the MiniSeq® (with Rapid reagents), instruments but in reverse-complement on patterned-flow-cell instruments such as iSeq®, MiniSeq® (with Standard reagents), NextSeq®, NovaSeq® 6000 and X series instruments.5
Troubleshooting After a Problem Run
Open Sequencing Analysis Viewer and plot Registration % and per-channel intensities for the index reads. A plunge in registration at cycle 1 or 2, especially if one channel’s intensity is flat, confirms imbalance. Re-pool with a PhiX spike-in or swap a few indices, then re-sequence. If the pool is needed regularly, rebuild it with the per-cycle counts described above.
Conclusion
Color balance may be invisible until it breaks, yet it governs whether a sequencer can keep its optical bearings through the index read. Four-channel systems were forgiving; two and one-channel chemistries are not. Even with brighter XLEAP® dyes, every cycle still needs light in every active channel. By relying on modern UDI plates, counting bases per cycle, and adding a diversified control, when necessary, laboratories can eliminate a common failure mode and protect both turnaround time and reagent budgets.
References:
- Illumina Index Adapters Pooling Guide (Document # 1000000041074 v13, July 2023).
- Illumina Knowledge Base: “Index color balancing for XLEAP SBS reagents on the NextSeq 1000/2000 and NovaSeq X/X Plus” (Article #8422, 2025).
- Illumina Support: “Illumina Unique Dual Indexes” (support-docs.illumina.com, accessed July 2025).
- Illumina Knowledge Base: “Troubleshooting running low diversity samples with low PhiX spike in on the iSeq 100” (Article #1369, 2024).
- Illumina Knowledge Base: “Guidelines for reverse complementing i5 sequences for demultiplexing” (Article #1800, 2025).


































