Y RNAs represent an intriguing class of small RNAs. Discovered as components of ribonucleoprotein complexes recognized by autoantibodies in systemic lupus erythematosus patients, Y RNAs were initially considered niche molecules restricted to autoimmune pathology1. However, subsequent work has revealed that they are essential in vertebrate systems for DNA replication, RNA surveillance, apoptosis and intercellular communication. Like other small RNAs, they are emerging as compelling biomarker candidates.
Human cells express four canonical Y RNAs which differ in length, abundance, and tissue distribution. They share a conserved stem–loop architecture and commonly bind the proteins Ro60 and La, yet each isoform exerts distinct biological roles. In addition to their full-length forms, Y RNAs are processed into specific fragments, similar to what has been described for tRNA. These Y RNA-derived fragments are stable in circulation and are increasingly studied. This review examines current knowledge on Y RNA biogenesis, isoform-specific biology, fragment classification, biomarker potential, and methods of detection.
Y RNA biogenesis, diversity, and functions
Y RNAs are transcribed by RNA polymerase III. Once synthesized, nascent Y RNAs are bound by the La protein, which stabilizes their 3′ ends and prevents premature degradation1,2. Association with the Ro60 protein follows, a process dependent on a conserved cytosine in the lower stem of the RNA. Ro60 binding modulates RNA stability and localization and also controls Ro60’s availability for misfolded RNA surveillance. Export of Y RNAs to the cytoplasm occurs predominantly via Exportin-5 in a RanGTP-dependent pathway, reminiscent of microRNA precursor trafficking3-6. Alternative routes have been reported.
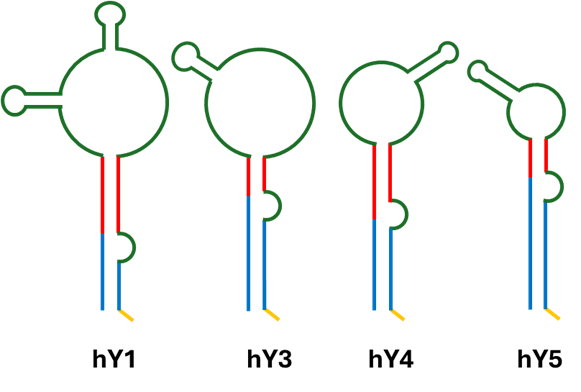
Although they share a conserve structure, the four canonical Y RNA isorforms distinct expression patterns and context-specific roles 7 (Figure 1).

Figure 1. Secondary structure of human Y RNA isoforms. Green portion corresponds to the loop domain, where cleavage of Y RNA takes place. Red corresponds to upper stem domain, crucial for DNA replication initiation. Blue is required for binding to Ro60 and yellow is needed for binding to La protein. Modified from Guglas et al (2020)8.
- hY1 RNA (~112 nt) is the most abundant isoform and is broadly expressed, with enrichment in proliferating epithelia and lymphoid tissues. It binds Ro60 with high affinity and plays a dual role: stabilizing Ro60 and regulating its surveillance activity, while also being indispensable for chromosomal DNA replication. Depletion of hY1 prevents S-phase entry in cultured human cells, establishing it as essential for cell proliferation in these systems9.
- hY3 RNA (~101 nt) is expressed at moderate levels across many tissues but shows notable enrichment in monocytes and activated T lymphocytes10. Like hY1, it contributes to replication initiation and RNA quality control but seems to be relevant in extracellular communication: hY3-derived fragments are abundant extracellular vesicle RNA profiles in plasma, making this isoform especially important in biomarker research11.
- hY4 RNA (~93 nt) is the least abundant isoform. While less characterized, hY4 appears to contribute context-specific regulatory functions. Fragments of hY4 are selectively exported in extracellular vesicles under certain conditions, suggesting a specialized role in immune modulation11-13.
- hY5 RNA (~83 nt) is the shortest isoform. Its expression is variable in normal tissues but appears elevated in some tumors, including lung, bladder, and cervical cancers. Functional knockdown of hY5 reduces proliferation and induces cytostasis, supporting its direct involvement in oncogenesis14. hY5 thus emerges as both a proliferation factor and a candidate cancer biomarker.
- Full-length Y RNAs can be processed into shorter molecules termed Y RNA-derived small RNAs (YsRNAs). These fragments are selectively generated, most prominently during apoptosis. During apoptosis, caspase-3, cleaves the RNA-binding protein PTBP1, relieving protection ofY RNAs and enabling their processing into small RNA species. This pathway is independent of the canonical microRNA pathway, as it does not involve Drosha or Dicer, enzymes essential for miRNA processing15,16.
At present, there is no standardized classification system for YsRNAs. However, studies have reported fragments of two predominant sizes (~24 nt and ~31 nt) and shown that most circulating YsRNAs derive from the 5′ ends of their parent, although 3’ end derived fragments have been reported13. This emerging evidence suggests that YsRNAs are structured products of regulated cleavage events, rather than by-products of RNA turnover.
Biomarker potential and detection of Y RNAs
The intrinsic properties of Y RNAs and their fragments, structural stability, evolutionary conservation, and enrichment in circulation make them compelling biomarker candidates. YsRNAs derived from hY3 and hY5 have been associated with systemic inflammation, autoimmunity, and cancer10,12. Their stability in serum and resistance to degradation further enhance their value in liquid biopsy applications. Full-length Y RNAs are also elevated in multiple tumors, with hY5 correlating with aggressive disease14.
Autoimmune conditions add another layer of relevance. Ro60-Y RNA complexes are direct targets of autoantibodies in systemic lupus erythematosus and Sjögren’s syndrome, linking Y RNA biology to disease mechanisms4. This duality as functional regulators and immunogenic complexes strengthens the case for their translational utility.
Y RNAs and YsRNAs can be detected using several methods. Quantitative RT-PCR remains a gold standard for isoform-specific quantification in tissues and fluids. However Small RNA sequencing
 NEXTFLEX Small RNA Sequencing Kit V4
is becoming more popular as it provides high-throughput profiling of full-length and fragmented Y RNAs, particularly in liquid biopsy. More recently, epitranscriptomic sequencing approaches promise to map modifications across Y RNAs and YsRNAs, potentially revealing new regulatory layers.
NEXTFLEX Small RNA Sequencing Kit V4
is becoming more popular as it provides high-throughput profiling of full-length and fragmented Y RNAs, particularly in liquid biopsy. More recently, epitranscriptomic sequencing approaches promise to map modifications across Y RNAs and YsRNAs, potentially revealing new regulatory layers.
Conclusions:
For the scientific community, Y RNAs highlight how small noncoding RNAs with initially niche associations can prove essential for fundamental cellular processes. For translational research, they present opportunities both as diagnostic tools and as potential therapeutic targets. As small RNA-sequencing
 NEXTFLEX Small RNA Sequencing Kit V4
technologies continue to advance, the coming decade will likely elevate Y RNAs and their fragments from biological curiosities to relevant players in RNA medicine.
NEXTFLEX Small RNA Sequencing Kit V4
technologies continue to advance, the coming decade will likely elevate Y RNAs and their fragments from biological curiosities to relevant players in RNA medicine.
References:
- Lerner, M.R., et al. (1981). Two Novel Classes of Small Ribonucleoproteins Detected by Antibodies Associated with Lupus Erythematosus. Science, 211,400-402. DOI:10.1126/science.6164096.
- Stefano, J.E. (1984). Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36(1):145–154. DOI: 10.1016/0092-8674(84)90083-7.
- Green, C.D., et al (1998). Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA, 4(7):750-65. doi: 10.1017/s1355838298971667.
- Stein, A.J., et al (2005). Structural Insights into RNA Quality Control: The Ro Autoantigen Binds Misfolded RNAs via Its Central Cavity. Cell, 121(4):529-539. DOI: 10.1016/j.cell.2005.03.009
- Sim, S., et al (2009). The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell. 20(5):1555-64. doi: 10.1091/mbc.e08-11-1094.
- Rutjers, S.A, et al (2001). Identification of a novel cis-acting RNA element involved in nuclear export of hY RNAs. RNA. 7(5):741-52. doi: 10.1017/s1355838201002503.
- Perreault, J., et al (2007). Ro-Associated Y RNAs in Metazoans: Evolution and Diversification, Molecular Biology and Evolution, 24(8):1678–1689, doi: 10.1093/molbev/msm084.
- Guglas, K., et al (2020). YRNAs and YRNA-Derived Fragments as New Players in Cancer Research and Their Potential Role in Diagnostics. Int. J. Mol. Sci. 21, 5682. Doi:10.3390/ijms21165682.
- Christov, C.P., et al (2006). Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol. 26(18):6993-7004. doi: 10.1128/MCB.01060-06.
- Estravís, M., et al (2022). RNY3 modulates cell proliferation and IL13 mRNA levels in a T lymphocyte model: a possible new epigenetic mechanism of IL-13 regulation. Journal of Physiology and Biochemistry. 79, 1-11. Doi: 10.1007/s13105-022-00920-6.
- Driedonks, T.A.P., et al. (2020). Y-RNA subtype ratios in plasma extracellular vesicles are cell type- specific and are candidate biomarkers for inflammatory diseases. J Extracell Vesicles. 9(1):1764213. doi: 10.1080/20013078.2020.1764213.
- Li C, et al (2022). Selective sorting and secretion of hY4 RNA fragments into extracellular vesicles mediated by methylated YBX1 to promote lung cancer progression. J Exp Clin Cancer Res. 41(1):136. doi: 10.1186/s13046-022-02346-w.
- Dhahbi, J.M., et al (2013). 5'-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics. 45(21):990-8. doi: 10.1152/physiolgenomics.00129.2013.
- Christov, C.P., et al (2008). Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br J Cancer. 11;98(5):981-8. doi: 10.1038/sj.bjc.6604254.
- Eskandari, E., Eaves, C.J. (2022). Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J Cell Biol. 221(6):e202201159. doi: 10.1083/jcb.202201159.
- Nicolas, F.E., et al (2012). Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Letters, 586(8):1226-1230. Doi: 10.1016/j.febslet.2012.03.026.


































