Transfer RNA (tRNA) is classically defined as the adaptor molecule bridging mRNA codons to amino acids during translation. In eukaryotes, mature tRNAs are typically 76–90 nts long, and each tRNA is bound to an amino acid by an aminoacyl tRNA synthetase before delivering its cognate amino acid to the ribosome.
Over the past decade, it has been shown that tRNAs play diverse non canonical roles. One exciting discovery is that mature and precursor tRNAs are specifically sheared into small fragments, generally called tRNA-derived fragments (tRFs) or tRNA halves (tiRNAs), with distinct regulatory functions. These fragments are not mere degradation products but are generated by regulated cleavage, interact with proteins or mRNA, and are implicated in cellular stress responses, epigenetic regulation, translation control, and disease states1-4.
Despite this progress, many aspects of their biogenesis, modification interplay, tissue specificity, and molecular mechanisms remain incompletely elucidated. In this blog we outline what we know and highlight critical open questions.
tRNA biogenesis and classification
Both cytoplasmic and mitochondrial tRNAs can produce fragments. Cleavage enzymes include Angiogenin, Dicer, RNase Z and RNase P. The determinants of tRNA cleavage specificity remain incompletely resolved.
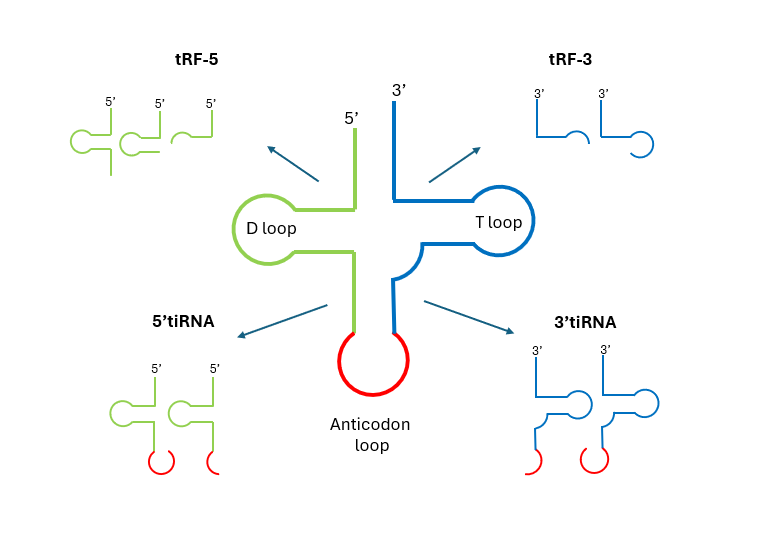
tRFs are ~14 -30 nucleotides in length. They are similar in length and have 5'-phosphate and 3'-hydroxyl groups like miRNA, for which reason they have attracted much attention over the years. tRFs are grouped into five major categories, four of which derive mature tRNA, while the last one originates from the UTR sequence of the precursor tRNAs5.
tRF 5, tRF 3 are originated by cleavage of the 5' and 3' ends of mature tRNAs, respectively. They are commonly found in small RNA data sets in all types of analysis, from yeast to human6 (Figure 1).

Figure 1. Main categories of tRFs and tiRNAs. Adapted from Xie et al (2020). Please notice there is no unified naming, and the different categories may appear under slightly different names in the literature.
tiRNAs have a length of ~30–40 nucleotides and are produced by cleaving across the anticodon loop under stress conditions and viral infections2. tiRNAs are classified in two groups according to whether the sequence contains the original 5' or 3' end (Figure 1).
Notably, full-length tRNAs, tRFs, and tiRNAs have been consistently detected in extracellular vesicles as well as in the cell-free RNA fraction of biofluids, indicating potential roles in intercellular communication and utility as minimally invasive biomarkers7.
Known or proposed tRNA functions
Translational regulation and stress response
Some tiRNAs inhibit global translation initiation by binding to eIF4G/A, promoting stress granule assembly. For example, angiogenin generated 5′ tiRNAs can displace eIF4F complexes, reducing cap dependent initiation under stress8,9. How broad is this mechanism across cell and tissue types is not yet understood. It is also unclear whether all stress conditions can generate tiRNAs, and what distinguishes functional species from background cleavage.
RNA interference
Several studies have reported that certain tRFs are loaded into Argonaute (AGO) complexes and can act like microRNAs, modulating target mRNA stability. The degree to which tRFs mimic miRNA targeting mechanism versus novel modes remains unclear.
For example, it has been shown displacement of YBX1 by tRFs suppresses oncogenic transcripts in breast cancer3. In human HEK293T cells, upregulating tRFs leads to the widespread repression of target genes. Rhizobial tRF-3 use host AGO1 to repress negative symbiotic regulators in the host. tRFs are also shown to hinder the replication of endogenous retroviruses by targeting its primer-binding site triggering immunity10.
Stem cells, development, fertility
Emerging work in mammalian stem cells and sperm maturation indicates that tRFs influence cell fate decisions and intergenerational information transfer11. The challenge remains to define what specific tRF species are necessary or sufficient in developmental fate decisions. Also, it is unclear how these fragments integrate within known epigenetic and signaling networks.
Link with tRNA modifications and network specificity
A major open frontier is how tRNA modifications (e.g. methylations, pseudouridine) influence tRF generation, stability, or function. Some fragments resist detection due to modification induced reverse transcription blockage. The impact of modifications on cleavage and how do these layers regulate fragment repertoires in physiology or disease remains largely uncharted.
Sequencing tRNAs and tRFs
Small RNA sequencing
 NEXTFLEX Small RNA Sequencing Kit V4
is the standard method to profile RNAs with length <200 nt, including tRNA, tRFs and tiRNAs. It enables high throughput discovery and quantification of species present in a sample at single-base resolution, although there a few challenges to consider.
NEXTFLEX Small RNA Sequencing Kit V4
is the standard method to profile RNAs with length <200 nt, including tRNA, tRFs and tiRNAs. It enables high throughput discovery and quantification of species present in a sample at single-base resolution, although there a few challenges to consider.
For example, many tRFs and tiRNAs (e.g. angiogenin generated), bear 5' OH or 2',3' cyclic phosphate ends which hinder standard adapter ligation. This can be overcome by treating the RNA upstream of library prep (e.g. T4 PNK or phosphatase). Protocols are readily available upon request.
Another challenge is the presence of internal modifications such as m¹A, m₃C, m¹G etc. They can block reverse transcriptase, causing drop outs and bias. Methods like PANDORA seq (which enzymatically demethylates before RT) have revealed previously under detected tRFs and tiRNAs12.
Finally, accurate alignment of tRNA-derived reads remains a significant bioinformatic challenge. Mature tRNAs often share high sequence similarity, sometimes differing by only one or two nucleotides. As a result, standard small RNA-seq alignment pipelines frequently struggle with multimapping, misassignment, or low confidence in read origin, particularly when dealing with short or modified fragments. To address these limitations, specialized tools such as tDRmapper, MINTmap, SPORTS1.0, and tsRNAsearch have been developed. These pipelines integrate curated tRNA annotation databases, consider both precursor and mature tRNA isoacceptors, and implement logic to account for CCA addition, mismatches due to modifications, and fragment ambiguity.
References:
- Yu, X., et al. (2021). tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics,11(1):461-469. doi: 10.7150/thno.51963.
- Li, S., et al. (2018). tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes (Basel), 9(5):246. doi: 10.3390/genes9050246.
- Fu, M., et al. (2023). Emerging roles of tRNA-derived fragments in cancer. Mol Cancer, 22, 30. doi: 10.1186/s12943-023-01739-5.
- Kumar, P., et al. (2016). Biogenesis and Function of Transfer RNA-Related Fragments. Trends Biochem. Sci., 41(8):679-689.
- Xie, Y., et al. (2020). Action mechanisms and research methods of tRNA-derived small RNAs. Sig. Transduct. Target Ther., 5, 109. doi: 10.1038/s41392-020-00217-4.
- Cole, C., et al. (2009). Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA, 15(12):2147-60. doi: 10.1261/rna.1738409.
- Tosar JP, Cayota, A. (2020). Extracellular tRNAs and tRNA-derived fragments. RNA Biol.17(8):1149-1167. doi: 10.1080/15476286.2020.1729584.
- Advani, V.M., Ivanov, P. (2019). Translational Control under Stress: Reshaping the Translatome. Bioessays, 41(5):e1900009. doi: 10.1002/bies.201900009.
- Ivanov, P., et al. (2011). Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell, 43(4):613-23. doi: 10.1016/j.molcel.2011.06.022.
- Kim, S.I., et al. (2025). A nuclear tRNA-derived fragment triggers immunity in Arabidopsis. Commun. Biol., 8, 533. doi:10.1038/s42003-025-07737-1.
- Guzzi, N., Bellodi, C,. (2020). Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol., 17(8):1214-1222. doi: 10.1080/15476286.2020.1732694.
- Shi, J., et al. (2021). PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol 23, 424–436. doi: 10.1038/s41556-021-00652-7.
For research use only. Not for use in diagnostic procedures.