Small nuclear RNAs (snRNAs) are short, non-coding RNAs of ~150 nucleotides that reside in the eukaryotic nucleus, where they associate with specific proteins to form small nuclear ribonucleoproteins (snRNPs). In humans, these complexes constitute the functional backbone of the spliceosome, a highly dynamic machine that excises introns and ligates exons during pre-mRNA processing1.
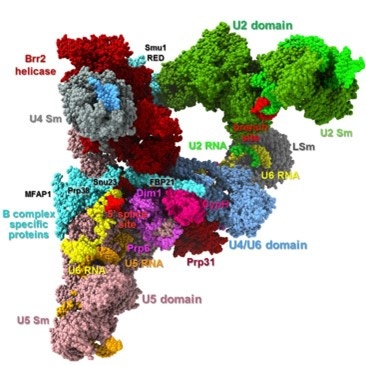
The major spliceosome, responsible for splicing > 99% of human introns, is built around the U1, U2, U4/U6, and U5 snRNPs, together with numerous auxiliary proteins that assemble in a stepwise manner on each intron (Figure 1). U1 snRNP recognizes the 5′ splice site, U2 binds to the branch point sequence, and the U4/U6·U5 tri-snRNP joins to form the precatalytic complex. During activation, U1 and U4 are displaced, allowing U6 and U2 to form the catalytic core that coordinates two magnesium ions required for the chemistry of splicing, while U5 aligns exons for ligation2,3 .
In parallel, the human genome encodes a minor spliceosome that acts on a small subset (<1%) of introns with distinct consensus sequences. This system uses U11, U12, U4atac, and U6atac snRNPs in combination with U54 .

Figure 1. Structure of pre-catalytic spliceosome. In humans (and other eukaryotes) the spliceosome does not exist as a single static machine. Instead, it goes through a series of assembly and activation intermediates, usually labeled E, A, B, B*, C, C* etc. The B spliceosome is one of these intermediates. From Bertram et al (2017)4.
Beyond their canonical role in splicing, snRNAs are known to contribute to other aspects of gene regulation. For example, 7SK snRNA regulates transcription elongation by sequestering the positive transcription elongation factor b (P-TEFb), while other snRNAs are implicated in nuclear organization and chromatin function3.
Splicing and classes of introns
Splicing is the process by which non-coding introns are removed from pre-mRNA and coding exons are ligated to form mature messenger RNA. It occurs through two sequential reactions. In the first step, the 2′ hydroxyl of the branch point adenosine attacks the 5′ splice site, generating a lariat intermediate. In the second step, the free 3′ hydroxyl of the upstream exon attacks the 3′ splice site, releasing the intron lariat and ligating the exons (Figure 2).
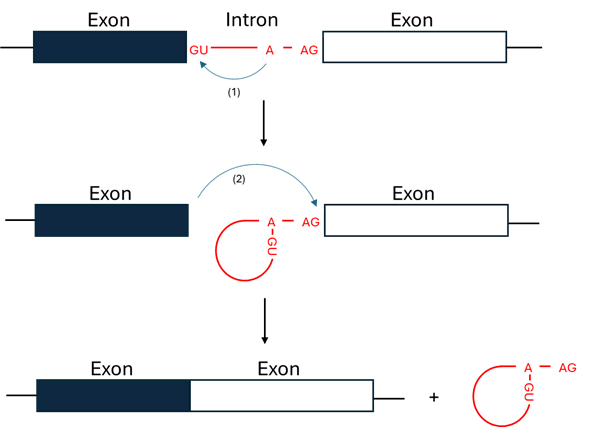
Figure 2. Diagram of RNA splicing
Introns are classified into two broad categories: U2-type introns, which make up more than 99% of human introns and are excised by the major spliceosome, and U12-type introns, which are rare (<1%) and processed by the minor spliceosome. These intron classes differ in their consensus sequences at the 5′ splice site, branch point, and 3′ splice site. U2-type introns typically have the canonical GU-AG boundaries, while U12-type introns feature GU-AG or occasionally AT-AC termini but with distinct internal consensus motifs that necessitate recognition by the specialized U11/U12-dependent spliceosome3,4.
| U2-type (major class) | U12-type (minor class) | |
|---|---|---|
| Spliceosome | Major spliceosome | Minor spliceosome |
| 5' splice site consensus | GU at the 5' end of intron | GU at 5' end, though some 5' ednds are AU with distinct internal motifs |
| 3′ splice site consensus | AG at intron end, preceded by polypyrimidine tract | AG at intron end, but usually lacks a polypyrimidine tract |
| Splicing efficiency | Generally fast and efficient | Generally slower and less efficient; more sensitive to mutations |
Table 1. Differences between the two classes of introns present in humans
This division into intron classes underscores why two distinct snRNA repertoires are required to support the dual spliceosomal systems operating in human cells.
Classification of snRNAs
Human snRNAs fall into two major classes with distinct biogenetic pathways and functional specializations (Table 2).
| Sm-class | Lsm-class | |
|---|---|---|
| Members | U1, U2, U4, U5, U7, U11, U12, U4atac | U6 and U6atac |
| Transcription | RNA polymerase II | RNA polymerase III |
| Cap structure | 2,2,7-trimethylguanosine cap | γ-monomethyl phosphate cap |
| Protein partners | Canonical Sm heptameric ring | Lsm2–8 protein ring |
| Sequence features | Conserved Sm site (AAUUUUUG), a single-stranded region flanked by stem-loops. | Conserved U-rich 3′ end |
| Location | Undergo nuclear export/import cycles | Nucleus |
Table 2. Differences between the two classes of snRNA in humans
Sm-class snRNAs, including U1, U2, U4, and U5, act primarily as recognition elements and structural scaffolds within the spliceosome. U1 initiates assembly by pairing with the 5′ splice site, U2 positions the branch point adenosine for nucleophilic attack, and U4–U6 interactions regulate activation of the catalytic center, while U5 stabilizes exon alignment. The minor spliceosome snRNAs (U11, U12, U4atac, and U6atac) perform analogous roles in U12-type intron splicing2.
In contrast, Lsm-class snRNAs, represented by U6 and U6atac, form the catalytic heart of the spliceosome. U6 base-pairs with U2 to create an RNA–RNA triplex that coordinates the catalytic magnesium ions, a function stabilized by the Lsm2–8 protein complex bound to its conserved uridine-rich 3′ tail. Unlike Sm-class snRNAs, U6 and U6atac are transcribed by RNA polymerase III, capped with a γ-monomethyl phosphate group, and remain nuclear throughout their life cycle. Perturbations of U6 or its Lsm-binding domain compromise splicing fidelity and disrupt global transcriptome integrity4,6.
Techniques to study snRNAs
The study of snRNAs has been transformed by NGS. Classical RNA-seq
 NEXTFLEX Rapid Directional RNA-Seq Kit 2.0
Discover
using as input total RNA where rRNA has been depleted
NEXTFLEX Rapid Directional RNA-Seq Kit 2.0
Discover
using as input total RNA where rRNA has been depleted
 NEXTFLEX RiboNaut rRNA Depletion Kit (Human / Mouse / Rat)
Discover
is particularly powerful. Please note that most snRNAs are not polyadenylated, and therefore they are usually underrepresented in poly(A)-enriched RNA-seq libraries. Classical RNA-seq provides information on full-length snRNAs and their connection to splicing dynamics and broader transcriptome context.
NEXTFLEX RiboNaut rRNA Depletion Kit (Human / Mouse / Rat)
Discover
is particularly powerful. Please note that most snRNAs are not polyadenylated, and therefore they are usually underrepresented in poly(A)-enriched RNA-seq libraries. Classical RNA-seq provides information on full-length snRNAs and their connection to splicing dynamics and broader transcriptome context.
Small RNA sequencing (small RNA-seq) offers a complementary perspective. This technique is designed to capture RNAs in the 20–200 nucleotide range. While small RNA-seq does not typically recover full-length snRNAs, it detects their processed fragments or degradation products. This is valuable not only for cataloguing RNA species but also for understanding turnover, processing, and potential fragment-based regulation, as has been shown in other small RNA species such as tRNA and Y RNA. Dedicated computational pipelines such as COMPSRA enable annotation of diverse small RNA biotypes within sequencing data, including snRNAs and snoRNAs6.
When both datasets are generated from the same biological samples, they create a multi-layered view of snRNA biology, combining quantitative expression profiles with fragmentomic signatures. This combined strategy can help distinguish changes in snRNA gene expression from alterations in processing or degradation, thereby enriching our understanding of nuclear RNA regulation.
References:
- Will CL, Lührmann R. (2011). Spliceosome structure and function. Cold Spring Harb Perspect Biol. 3(7):a003707. doi: 10.1101/cshperspect.a003707.
- Matera AG, Wang Z. (2014) A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 15(2):108-21. doi: 10.1038/nrm3742. Erratum in: Nat Rev Mol Cell Biol. 15(4):294.
- Patel AA, Steitz JA. (2003). Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 4(12):960-70. doi: 10.1038/nrm1259.
- Bertram, K., et al. (2017). Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation. Cell, (170):4, 701 - 713.e11. doi: 10.1016/j.cell.2017.07.011
- Didychuk, A.L., et al. (2018). The life of U6 small nuclear RNA, from cradle to grave. RNA.24(4):437-460. doi: 10.1261/rna.065136.117.
- Li, J., et al. (2020). COMPSRA: a COMprehensive Platform for Small RNA-Seq data Analysis. Sci Rep 10, 4552. doi:10.1038/s41598-020-61495-0.


































