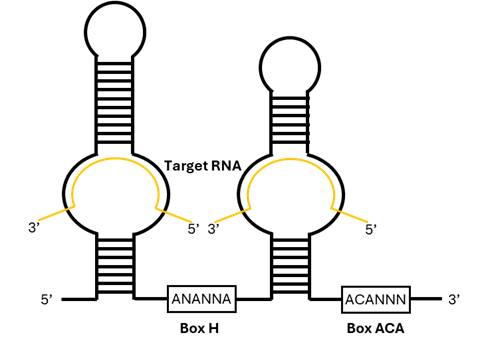
Small nucleolar RNAs (snoRNAs) are a conserved and abundant type of non-coding RNAs present in all eukaryotes sequenced so far. With 60–300 nucleotides in length, snoRNAs were first recognized for their location in the nucleolus and their essential role in ribosome biogenesis. They associate with conserved proteins to form small nucleolar ribonucleoproteins (snoRNPs), guiding chemical modifications of ribosomal RNAs and certain small nuclear RNAs (Figure 1)1.

Figure 1. Scheme of an H/ACA snoRNA bound to its targets. Modified from Bergeron et al (2023)2.
For many years, snoRNAs were perceived as “housekeeping” RNAs, performing their modification duties in the background of the cell. Over the last decade, however, this view has changed. Beyond their canonical functions, snoRNAs have been implicated in pre-rRNA processing, splicing regulation, mRNA modification, chromatin organization, and even in generating smaller RNA fragments that behave like microRNAs.
Genetic evidence now links snoRNA mutations or deletions to human diseases ranging from rare neurodevelopmental syndromes to cancer3,4. With this expanded appreciation comes a renewed interest in how to detect, quantify, and functionally analyze snoRNAs using next-generation sequencing (NGS) technologies.
Biogenesis and assembly of snoRNPs
Most vertebrate snoRNAs are encoded within introns of protein-coding or non-coding genes transcribed by RNA polymerase II. Following transcription, introns are excised during splicing, and the snoRNA sequence is liberated from the debranched lariat. Exonucleolytic trimming then produces a mature snoRNA, which assembles into a snoRNP particle5,6. This intronic arrangement tightly couples snoRNA expression to the transcriptional program of their host genes.
Box C/D snoRNAs are defined by conserved C (RUGAUGA) and D (CUGA) sequence motifs that fold into a characteristic kink-turn (K-turn) structural element. This architecture is specifically recognized by the core proteins fibrillarin, NOP56, NOP58, and SNU13, which together form the catalytically competent C/D snoRNP7,8. By contrast, H/ACA snoRNAs adopt a bipartite hairpin–hinge–hairpin–tail secondary structure and terminate with an ACA trinucleotide at their 3′ end. They assemble with a distinct set of core proteins: dyskerin, NHP2, NOP10, and GAR1, to generate functional H/ACA snoRNPs6.
Although box C/D and H/ACA snoRNAs represent the two widely accepted categories, additional specialized subfamilies exist. These include chromatin-associated RNAs (scaRNAs), which are functionally and structurally related but distinct in localization and substrate specificity, and a small number of “orphan” snoRNAs that lack clearly defined targets6,8. Importantly, not all snoRNAs act as modification guides. Members such as U3, U8, and snR30 are essential for pre-rRNA cleavage and ribosomal subunit maturation, highlighting the functional diversity within the snoRNA repertoire7,9.
Beyond canonical modifications of rRNA
The canonical role of snoRNAs is to guide chemical modifications (2′-O-methylation and pseudouridylation) at hundreds of conserved positions in rRNA. These modifications fine-tune ribosome function by stabilizing local RNA structure, shaping the peptidyl transferase center, decoding sites, and intersubunit bridges. Knockouts of specific snoRNAs or their associated enzymes frequently impair ribosome assembly, reduce translational accuracy, or alter the ribosome’s responsiveness to stress10.
Yet snoRNAs also perform non-canonical activities. Some regulate alternative splicing by masking splice sites or modulating local RNA structure. Others generate snoRNA-derived RNAs (sdRNAs) that can act in a miRNA-like manner, associating with Argonaute proteins to direct post-transcriptional gene silencing11. Emerging evidence even links snoRNAs to chromatin modifications and transcriptional gene regulation12.
Profiling snoRNAs with RNA-seq
Historically, snoRNAs were studied by northern blotting, primer extension, and RNase protection assays. While informative, these approaches were low-throughput, labor-intensive, and biased toward abundant species.
NGS has transformed snoRNA research. However, snoRNAs present technical challenges: their structured nature, intronic origins, and lack of poly(A) tails mean they are often under-represented in conventional mRNA-seq. The most straightforward approach to overcome these problems is to perform total RNA-seq with rRNA depletion.
This captures intronic and mature snoRNAs, though some library bias remains. In parallel, Small RNA-seq (designed for miRNAs) can be used to capture the snoRNA-derived fragments (~18-30 nt) that behave like miRNAs, making it a valuable tool for studying their expression patterns, biogenesis, and potential regulatory roles. Terminal chemistries can reduce ligation efficiency (e.g., 2′,3′-cyclic phosphate; 5′-OH). We recommend a PNK end-repair treatment before starting library prep to improve recovery. Dedicated computational resources now complement these methods. Databases such as snoDB 2.0 and snOPY curate guide–target interactions and orthology relationships, while pipelines like COMPSRA annotate snoRNAs alongside other small RNAs12
Future outlook
SnoRNAs have emerged from the shadows of “housekeeping” biology into the spotlight of modern RNA research. They exemplify how a seemingly narrow set of non-coding RNAs can impact fundamental processes from ribosome assembly to gene regulation and disease. Technological innovations now allow researchers to move beyond cataloging individual snoRNAs toward profiling entire “snoRNAomes” and connecting them to functional outputs.
Looking forward, several areas seem poised for rapid progress: the discovery of snoRNA-derived small RNAs with regulatory functions; the integration of modification mapping with ribosome profiling to uncover how rRNA chemical diversity shapes translation; and the exploration of snoRNAs as diagnostic biomarkers or therapeutic targets. As datasets expand and sequencing technologies mature, snoRNAs are unlikely to remain “small” players in RNA biology for much longer.
References:
- Bergeron, D., et al. (2021). SnoRNA copy regulation affects family size, genomic location and family abundance levels. BMC Genomics 22, 414. doi:10.1186/s12864-021-07757-1.
- Bergeron, D., et al. (2023). snoDB 2.0: an enhanced interactive database, specializing in human snoRNAs. Nucleic Acids Research,51(D1), D291–D296. doi:10.1093/nar/gkac835.
- Jenkinson, E.M., et al. (2016)- Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat Genet. 2016 Oct;48(10):1185-92. doi: 10.1038/ng.3661. Erratum in: Nat Genet. 2017 Jan 31;49(2):317. doi: 10.1038/ng0217-317b.
- Gawade K, Raczynska KD. Imprinted small nucleolar RNAs: Missing link in development and disease? Wiley Interdiscip Rev RNA. 2023 Sep 18:e1818. doi: 10.1002/wrna.1818. Epub ahead of print. PMID: 37722601.
- Hesselberth, J.R. (2013). Lives that introns lead after splicing. Wiley Interdiscip Rev RNA. 4(6):677-91. doi: 10.1002/wrna.1187.
- Massenet, S. et al (2017). Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017 14(6):680-692. doi: 10.1080/15476286.2016.1243646.
- Kiss, T. (2002). Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 109(2):145-8. doi: 10.1016/s0092-8674(02)00718-3.
- Watkins, N.J., Bohnsack M.T. (2012). The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 3(3):397-414. doi: 10.1002/wrna.117.
- Henras, A.K., et al (2008). The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 65(15):2334-59. doi: 10.1007/s00018-008-8027-0.
- King, T.H., et al (2003). Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 11(2):425-35. doi: 10.1016/s1097-2765(03)00040-6.
- Ender, C., et al (2008). A human snoRNA with microRNA-like functions. Mol Cell, 32(4):519-28. doi: 10.1016/j.molcel.2008.10.017.
- Bratkovič, T., et al (2020). Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 48(4):1627-1651. doi: 10.1093/nar/gkz1140.
- Li, J., et al. (2020). COMPSRA: a COMprehensive Platform for Small RNA-Seq data Analysis. Sci Rep 10, 4552. doi:10.1038/s41598-020-61495-0.


































