
NEXTFLEX Cell Free DNA-Seq Library Prep Kit 2.0



| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | DNAシーケンス |



Product information
Overview
By simplifying multi-step cfDNA library prep into a robust, streamlined and automation-friendly workflow, the NEXTFLEX Cell Free DNA-Seq Kit 2.0 removes the usual throughput, and indexing bottlenecks and reduces the bias that limit liquid-biopsy, prenatal, and transplant-monitoring studies. The benefits below highlight how the NEXTFLEX Cell Free DNA-Seq Library Prep Kit 2. conserves precious plasma or serum input, scales from pilot projects to population studies, and plugs seamlessly into Illumina® and Element® sequencing pipelines.
- Low input: robust performance from 10 ng cfDNA, conserving precious liquid-biopsy samples
- Rapid turnaround: two-hour total workflow with minimal hands-on time accelerates time-to-result
- High library complexity: optimized ligation chemistry boosts unique read counts and reduces duplicates
- Minimal bias: uniform genome coverage improves sensitivity for SNVs, indels, CNVs, and aneuploidies
- Flexible multiplexing: 1,536 UDIs or 96 UDI-UMIs combat index hopping and enable error correction
- Automation-ready: validated protocols for Revvity liquid-handlers support scalable clinical-research pipelines
- Platform versatility: proven on Illumina® and Element Biosciences® sequencers for broad lab adoption
Additional product information
Low-Input cfDNA Performance
Generate deep-coverage libraries from just 10 ng of plasma- or serum-derived cfDNA, the amount typically recovered from under 3 mL samples. Our chemistry preserves the native cfDNA fragment profile, which presents 5′-phosphate and 3′-hydroxyl ends typical of nuclease cleavage. That means you can profile early-stage cancers or run cell-free fetal DNA tests on these small input samples without resorting to pre-amplification or whole-genome amplification that can skew variant calls.

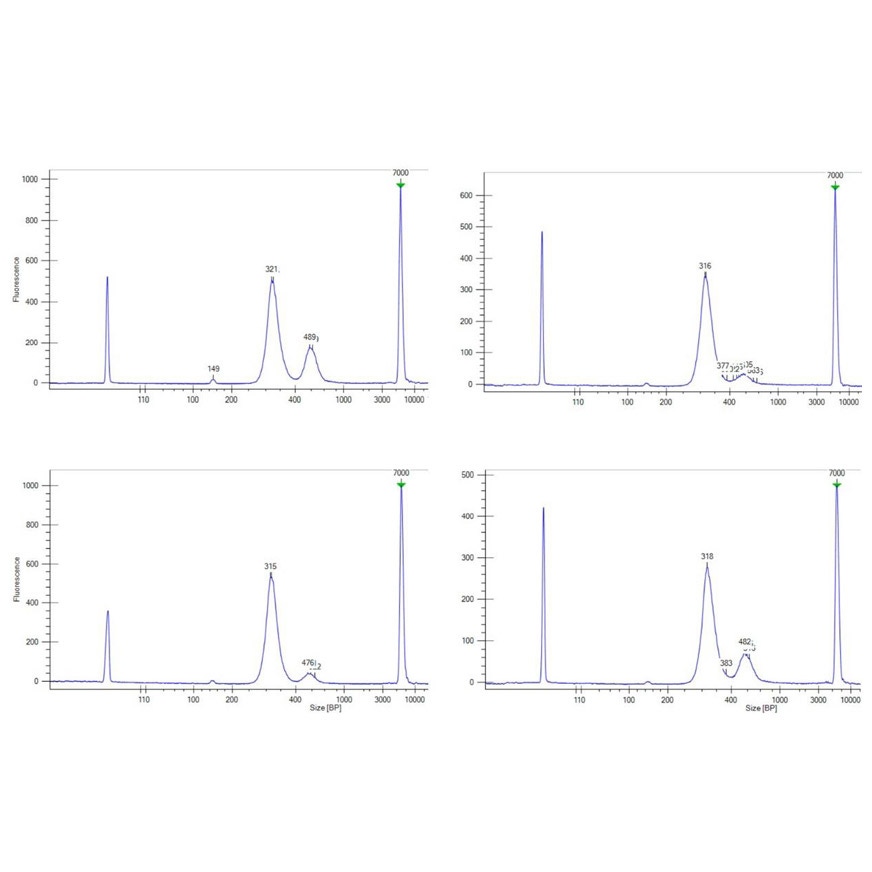
Figure 1: Electropherograms of cfDNA libraries prepared from 10 ng plasma cfDNA with the NEXTFLEX Cell Free DNA-Seq Kit 2.0 show a clean ~320 bp peak, negligible adapter-dimer, and highly consistent low-input performance.
Accelerated cfDNA Workflow Efficiency
A consolidated, single-tube end-repair/A-tailing step merges multiple enzymatic reactions, while magnetic-bead clean-ups are tuned for fast binding and elution. The result is a roughly 2-hour start-to-finish protocol with minimal hands-on pipetting, short enough to move from sample intake to sequencer load in the same shift. Less bench time frees staff for higher value tasks, reduces pipetting error, and helps labs hit aggressive turnaround targets.
High-Complexity Libraries with Uniform Genome Coverage
Optimized ligation chemistry and precisely calibrated adapter stoichiometry maximize single-insert tagging efficiency, minimizing adapter-dimer formation and PCR duplicates while enriching the pool of unique cfDNA molecules that reach the flow cell. At the same time, a balanced buffer matrix, tuned for salt, PEG, and enzyme concentrations, suppresses sequence-dependent bias, delivering uniform coverage across GC-rich, AT-rich, and repetitive regions from telomere to telomere. Together, the resulting high library complexity and even genome representation drive deeper usable read depth and sharper quantitative resolution, enabling confident detection of low-frequency SNVs, indels, CNVs, and aneuploidies, essential for minimal-residual-disease monitoring, clonal evolution analysis, and other applications that demand analytical sensitivity.
Scalable Multiplexing & Error Correction
Select from 1,536 NEXTFLEX Unique Dual Index (UDI) pairs to pool thousands of samples per flow cell with negligible index crosstalk or deploy the 96-plex NEXTFLEX® UDI-UMI set when single-molecule error correction is needed for ultra-rare variant or MRD applications. Both index formats slot seamlessly into Illumina® and Element Biosciences® workflows, so you can dial in either throughput or precision without altering the core protocol.
Balanced cfDNA pools without extra QC steps
For high-plex cfDNA studies, apply NEXTFLEX NGS Library Normalization Beads post-PCR to equalize library molarity and pool directly, saving time and reducing variability in per-sample reads. On-bead normalization preserves the native cfDNA fragment profile and supports consistent, deep coverage at scale.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Product Group |
DNA-seq
|
| Shipping Conditions |
Dual Temperature
|
| Unit Size |
8 rxns
|
Citations
- Raman L, Van der Linden M, De Vriendt C, et al. (2022). Shallow-depth sequencing of cell-free DNA for Hodgkin and diffuse large B-cell lymphoma (differential) diagnosis: a standardized approach with underappreciated potential. Haematologica, 107(1), 211–220. https://doi.org/10.3324/haematol.2020.268813
- Zviran A, Schulman RC, Shah M, et al. (2020). Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nature Medicine, 26, 1114–1124. https://doi.org/10.1038/s41591-020-0965-7
- Brenner T, Decker SO, Vainshtein Y, et al (2025). Improved pathogen identification in sepsis or septic shock by clinical metagenomic sequencing. J Infect. 91(3):106565. https://doi.org/ 10.1016/j.jinf.2025.106565
- Müller J, Hartwig C, Sonntag M, et al. (2024). A novel approach for in vivo DNA footprinting using short double-stranded cell-free DNA from plasma. Genome Research, 34(8), 1185–1195.
- Mauger F, Horgues C, Pierre-Jean M, et al. (2020). Comparison of commercially available whole-genome sequencing kits for variant detection in circulating cell-free DNA. Scientific Reports, 10, 6190. https://doi.org/10.1038/s41598-020-63102-8
FAQs
-
What is the minimum cfDNA input I can use?
-
Which sample types work best?
-
How long does the workflow take, and how much hands-on time will I need?
-
Can I automate the protocol?
-
Which sequencers are supported?
-
What library size profile should I expect?
-
How do NEXTFLEX UDI-UMI adapters improve data quality?
-
Are the libraries compatible with hybrid-capture panels or shallow WGS?
Resources
Are you looking for resources, click on the resource type to explore further.
Benefit of an automated cfDNA analysis workflow
The presence of double stranded, circulating cell free DNA (cfDNA) in blood plasma...
Read the Mimix brochure to explore our range of cell line-derived Mimix reference standards which provide more patient-like...


How can we help you?
We are here to answer your questions.






























