
NEXTFLEX NGS Library Normalization Beads



| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | NGS Accessory |
For research use only. Not for use in therapeutic or diagnostic procedures.



Product information
Overview
NEXTFLEX™ NGS Library Normalization Beads capture a defined amount of library DNA during final cleanup to standardize input, promoting uniform cluster formation and balanced read counts across samples.
- On-bead library normalization that equalizes sample molarity.
- Eliminates separate quantitation and manual pooling, reducing hands-on time and turnaround.
- Saves ~3 hours per 96-sample batch in high-throughput workflows.
- Optimized protocol reduces variability that can lead to over- or under-clustering.
- Can be seamlessly integrated into most library preparation protocols.
- Eliminates the capital expense of buying an instrument for normalization.
- Easily automated protocol can further reduce hands-on time.
- Not for use with PCR-free workflows.
Additional product information
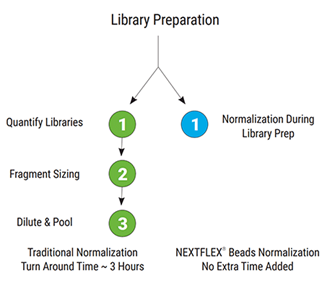
On-bead normalization with NEXTFLEX Library Normalization Beads
These beads capture a defined mass of library DNA so NGS libraries exit the workflow at comparable molarity. This removes post-library qPCR or fluorometric quantitation and manual pool balancing, cutting hands-on time and consumables. Labs typically save about three hours per 96-sample batch. On-bead normalization also reduces sample-to-sample variability, helping avoid over- or under-clustering and the resequencing that can follow, with no extra quantitation reagents or instruments required.

Figure 1: Workflow comparison of post-library normalization vs on-bead normalization with NEXTFLEX Library Normalization Beads.
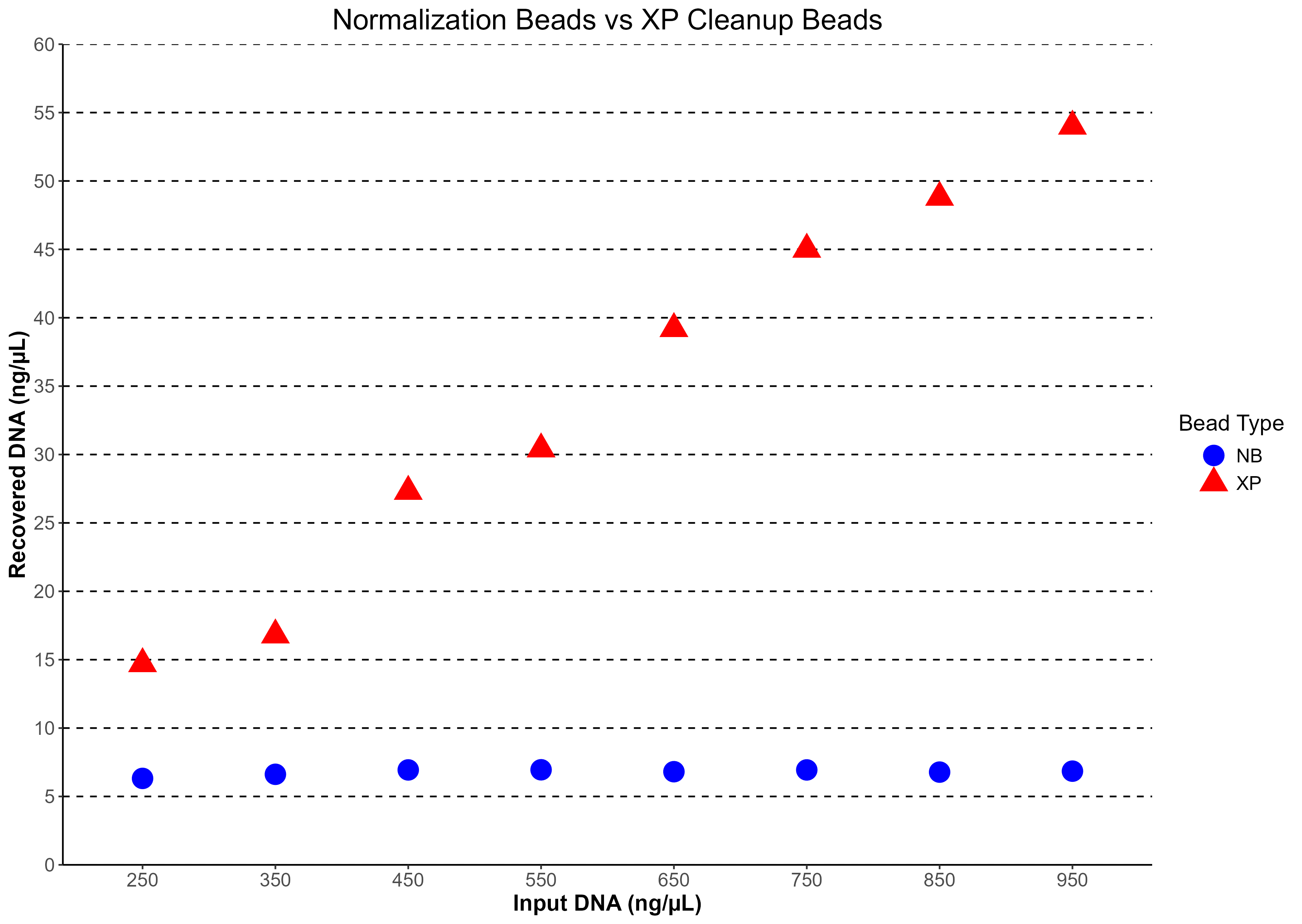
Figure 2: Normalization beads maintain an approximately constant recovered DNA concentration across a wide range of input DNA, demonstrating true normalization. In contrast, SPRI cleanup scales with input, increasing recovered DNA over the same range.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Product Group |
NGS Accessory
|
| Shipping Conditions |
Shipped Ambient
|
| Unit Size |
96 rxns
|
FAQs
-
What do Normalization Beads do?
-
When should I add the beads to my workflow?
-
Which library types are compatible?
-
Are the beads platform-agnostic?
-
What input concentration range works best?
-
What normalized output should I expect?
-
Can I pool directly after normalization?
-
Do the beads affect size distribution or library complexity?
-
Do I need additional reagents or special equipment?
Resources
Are you looking for resources, click on the resource type to explore further.


How can we help you?
We are here to answer your questions.






























