
NEXTFLEX 16S V4 Amplicon-Seq Kit 2.0



The NEXTFLEX™ 16S V4 Amplicon-Seq Kit 2.0 prepares sequencing libraries that amplify the V4 hypervariable region of the 16S rRNA gene for microbiome profiling. The two-step PCR workflow with minimal cleanups maximizes recovery and produces multiplexed amplicon libraries that do not require custom sequencing primers. Libraries are compatible with paired-end 2×300 sequencing on Illumina® MiSeq® and Element® Aviti® sequencers, using overlapping reads across V4 for accurate taxonomic assignment. Typical input is 1–50 ng of high-quality genomic DNA, enabling a fast, reliable amplicon sequencing workflow for bacterial community profiling. Choosing the V4 region provides broad, well-characterized bacterial coverage with the widely used 515F/806R primers. The resulting overlapping 2×300 reads support accurate amplicon sequence variant (ASV) calling and are highly compatible with established reference databases and analysis pipelines.
The kits include up to 384 barcoded PCR II primers to improve the efficiency of your workflow through high throughput indexing and pooling. In the product listing, the “S” variants include access to 16S analysis through Cosmos-Hub, which runs a DADA2-based 16S pipeline with SILVA taxonomy to generate quality-controlled ASV tables and interactive visualizations.
| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | 16Sアンプリコンシーケンス |
The NEXTFLEX™ 16S V4 Amplicon-Seq Kit 2.0 prepares sequencing libraries that amplify the V4 hypervariable region of the 16S rRNA gene for microbiome profiling. The two-step PCR workflow with minimal cleanups maximizes recovery and produces multiplexed amplicon libraries that do not require custom sequencing primers. Libraries are compatible with paired-end 2×300 sequencing on Illumina® MiSeq® and Element® Aviti® sequencers, using overlapping reads across V4 for accurate taxonomic assignment. Typical input is 1–50 ng of high-quality genomic DNA, enabling a fast, reliable amplicon sequencing workflow for bacterial community profiling. Choosing the V4 region provides broad, well-characterized bacterial coverage with the widely used 515F/806R primers. The resulting overlapping 2×300 reads support accurate amplicon sequence variant (ASV) calling and are highly compatible with established reference databases and analysis pipelines.
The kits include up to 384 barcoded PCR II primers to improve the efficiency of your workflow through high throughput indexing and pooling. In the product listing, the “S” variants include access to 16S analysis through Cosmos-Hub, which runs a DADA2-based 16S pipeline with SILVA taxonomy to generate quality-controlled ASV tables and interactive visualizations.



Product information
Overview
- Straightforward workflow: two PCR steps with minimal cleanups; no custom sequencing primers required.
- Cleanup beads included: NEXTFLEX NGS Cleanup Beads are supplied with the kit, allowing PCR cleanups without purchasing separate beads. This reduces consumables cost and simplifies purchasing.
- Accurate taxonomic assignment: overlapping paired-end 2×300 reads across the V4 region on Illumina® MiSeq® and Element® Aviti® sequencers support robust error correction and ASV recovery.
- Flexible Input: validated conditions for 1–50 ng genomic DNA help maintain consistency across samples.
- Multiplexing Options: from 12 to 384 PCRII Barcodes are available to support projects of all sizes.
- Analysis: product variants with “S” in their SKU include Cosmos-Hub 16S analysis (DADA2 with SILVA) to generate quality-controlled ASV tables and interactive reports.
- Automation: protocols are available for Sciclone™ NGS, NGSx, and Sciclone G3 NGSx iQ™ Workstations to automate 16S sequencing.
Use this kit when you want a dependable 16S V4 library prep that fits standard 2×300 sequencing on Illumina® MiSeq® or Element® Aviti® sequencers and hands off cleanly to downstream taxonomic analysis, including the Cosmos-Hub.
Additional product information
Accurate taxonomic assignment with full 16S V4 overlap and flexible input
Paired-end 2×300 sequencing fully covers the V4 insert, so forward and reverse reads overlap across every base. That overlap enables pair-merging, model-based error correction, and chimera removal, which improves discrimination among closely related bacteria and reduces misclassification in downstream taxonomic calls.

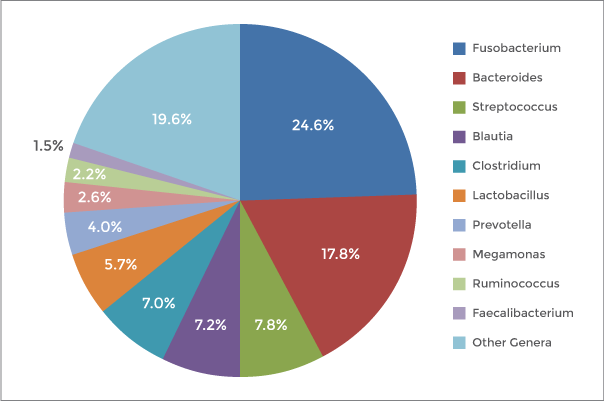
Figure 1: Genus-level composition from 16S V4 libraries prepared with the NEXTFLEX 16S V4 Amplicon-Seq Kit 2.0.
Accurate assignment matters because it drives the conclusions you report: community composition, relative abundance, and changes across conditions. Using exact sequence variants rather than broad clusters increases resolution and comparability across studies, which is especially useful for longitudinal work, method validation, and meta-analysis.
The kit supports a practical input range of 1–50 ng genomic DNA, so typical extractions can go straight into PCR without additional concentration. To maintain consistency across that range, the protocol provides cycle-number guidance that is adjusted to input amount, which helps achieve sufficient yield while limiting over-amplification artifacts.
Not sure when to choose 16S over metagenomics for microbial research? Read a quick comparison that outlines the tradeoffs.
Cosmos-Hub 16S analysis (included with “S” variants)
Product variants above with “S” in their SKU number include access to Cosmos-Hub, an online analysis platform that moves your 16S V4 FASTQ files to interpretable results with a standardized, version-controlled workflow, enabling fast and easy analysis of complex microbiome data. You receive a quality-controlled ASV table with taxonomy, clear QC summaries, and publication-ready visualizations. Comparative analysis is built in, including tables, heatmaps, bar and stacked plots, multiple alpha and beta diversity indices, abundance distribution views, differential abundance testing, and statistics between groups. Exports are provided as tab-separated tables and PDFs, with options to download individual charts as PNG or SVG files for downstream use. Learn more about Cosmos-Hub.


Revvity 16S rRNA analysis – online demonstration
Now you can schedule a one-on-one 30-minute online demonstration that will cover everything you need to know about the Cosmos-Hub® which is now available with Revvity’s NEXTFLEX™ 16S rRNA Amplicon-seq Kits. You will be able to see the analysis demonstrated and our experts will be able to answer any questions that you may have. Please choose a timeframe that suits you best and we will provide you a Microsoft Teams personal link to join when the time comes. We look forward to speaking with you.
Automation-ready
Automation protocols are available for the Sciclone NGS and NGSx workstations, supporting walkaway NGS library preparation and higher throughput in 96-well plates while reducing manual steps and variability. The Sciclone G3 NGSx iQ workstation adds an on-deck thermal cycler to automate PCR steps end to end. Together, these options standardize results and streamline batching without changing the kit chemistry.
Choose the 16S region that fits your study
Revvity offers 16S library prep kits for V4, V3–V4, and V1–V3 so you can align region choice with your objectives: use V4 when you want the shortest and most widely adopted amplicon that fits common read lengths and large legacy datasets, choose V3–V4 when added phylogenetic signal across two adjacent hypervariable domains will help resolution, or select V1–V3 when your assay or consortium specifies that region or certain clades are best distinguished there.
Specifications
| Analysis |
Not Included
|
|---|---|
| Automation Compatible |
Yes
|
| Barcodes |
1 - 96
|
| Product Group |
16S amplicon-seq
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
192 rxns
|
References
- Luk, B., Veeraragavan, S., Engevik, M., Balderas, M., Major, A., Runge, J., . . . Versalovic, J. (2018). Postnatal colonization with human “infant-type” Bifidobacterium species alters behavior of adult gnotobiotic mice. Plos One, 13(5). doi:10.1371/journal.pone.0196510.
- Moon, C., Stupp, G. S., Su, A. I., & Wolan, D. W. (2017). Metaproteomics of colonic microbiota unveils discrete protein functions among colitic mice and control groups. doi:10.1101/219782.
- Stalenhoef, J. E., et al. (2017). Fecal Microbiota Transfer for Multidrug-Resistant Gram-Negatives: A Clinical Success Combined With Microbiological Failure. Open Forum Infectious Diseases, 4(2). doi:10.1093/ofid/ofx047
- Whon TW, Chung WH, Lim MY, et al. (2018). The effects of sequencing platforms on phylogenetic resolution in 16S rRNA gene profiling of human feces. Scientific Data, 5:180068. doi:10.1038/sdata.2018.68
- Jovel J, Patterson J, Wang W, et al. (2016). Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front Microbiol, 7:459. doi:10.3389/fmicb.2016.00459.
- Long J, Cai Q, Steinwandel M, et al. (2017). Association of oral microbiome with Type 2 diabetes risk. J Periodontal Res, 52(3):636-643. doi:10.1111/jre.12432.
- Yang Y, Cai Q, Zheng W, et al. (2019). Oral microbiome and obesity in a large study of low-income and African-American populations. J Oral Microbiol, 11(1):1650597. doi:10.1080/20002297.2019.1650597.
- Curry KD, Wang L, Marcott PF, et al. (2022). Emu: species-level microbial community profiling of full-length 16S rRNA Oxford Nanopore sequencing data. Nat Methods, 19:845-853.
- Kartal E, Nitschke J, Zhernakova A, et al. (2022). A faecal microbiota signature with high specificity for pancreatic cancer. Gut, 71:1359–*.
- Yang CJ, Song JS, Yoo JJ, et al. (2024). 16S rRNA Next-Generation Sequencing May Not Be Useful for Examining Suspected Cases of Spontaneous Bacterial Peritonitis. Medicina, 60:289. doi:10.3390/medicina60020289.
FAQs
-
What sequencing platforms and read lengths are supported?
-
What DNA input range does the kit support?
-
Do I need to buy separate cleanup beads?
-
Which indexes are included, and can I scale higher multiplexing?
-
Which primers are used to target the V4 region?
-
Can I use this kit for archaeal profiling?
-
Why is paired-end 2×300 important for this V4 kit?
-
What do the “S” variants include for data analysis?
-
Is the workflow compatible with automation?
Resources
Are you looking for resources, click on the resource type to explore further.
Metagenomics holds tremendous promise, yet it is not without its constraints, especially when it comes to the scarcity of powerful...


How can we help you?
We are here to answer your questions.






























