
NEXTFLEX 16S V3 – V4 Amplicon-Seq Library Prep Kit



| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | 16Sアンプリコンシーケンス |



Product information
Overview
- Straightforward workflow: two PCR steps with minimal cleanups; no custom sequencing primers required.
- Beads included: NEXTFLEX NGS Cleanup Beads come in the kit, so PCR cleanups are covered without a separate purchase, which can reduce consumables spend and simplify ordering.
- Confident taxonomy calls: overlapping paired-end 2×300 reads across the V3-V4 amplicon on Illumina MiSeq and Element Aviti enable strong error correction and ASV recovery.
- Flexible Input: validated conditions for 1 – 50 ng genomic DNA help maintain consistency across samples.
- Scalable indexing: choose from 48 to 96 PCR II barcodes (384 available upon request) to match project size.
- Built-in analysis option: SKUs with an “S” include Cosmos-Hub 16S analysis using DADA2 with SILVA, producing quality-controlled ASV tables and interactive reports.
- Automation ready: methods available for Sciclone NGS and NGSx workstations, including G3 NGSx iQ.
Use this kit when you need reliable 16S V3-V4 library prep for standard 2×300 paired-end runs on Illumina MiSeq or Element Aviti, with a clean handoff to downstream analysis in Cosmos-Hub.
Additional product information
Simple 16S V3–V4 workflow, beads included, flexible multiplexing
The kit uses a two-PCR design. PCR I generates the V3–V4 amplicon from genomic DNA, then a bead cleanup removes primers and small fragments. PCR II adds the barcoded PCR II primers for indexing, followed by a final cleanup and pooling.
NEXTFLEX NGS Cleanup Beads are supplied with the kit. This helps reduce variability and avoids separate consumables purchase.
Indexing scales with your study: choose 48 or 96 barcode sets, with expanded options available up to 384 unique PCR II barcodes (upon request). Each barcode includes two reactions.
Accurate V3–V4 taxonomic assignment with overlapping reads and flexible input
Paired-end 2×300 sequencing spans the ~610 bp V3–V4 library and yields overlapping forward and reverse reads. That overlap enables read merging, quality-aware error modeling, and chimera filtering, which improves separation of closely related taxa and reduces misclassification in downstream calls.

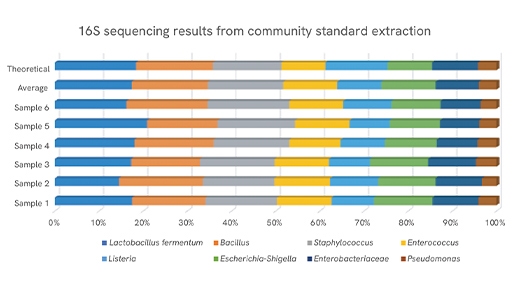
Figure 1: Population results displayed as the percentage of total 16S DNA present after sequencing the extracted Zymo Community Standard
Accurate assignment matters because it shapes the conclusions you report, including community composition, relative abundance, and changes across conditions. Using exact sequence variants instead of broad clusters improves resolution and comparability across studies, which is especially helpful for longitudinal designs, method validation, and meta-analysis.
The kit supports a practical input of 1–50 ng DNA, so typical extracts can go straight into PCR without extra concentration. To keep results consistent across that range, the protocol provides cycle-number guidance matched to input, which helps achieve sufficient yield while limiting over-amplification artifacts.
Not sure when to choose 16S or metagenomics for microbial work? Read a short comparison that outlines the tradeoffs.
Cosmos-Hub 16S analysis (included with “S” variants)
Product variants above with “S” in their SKU number include access to Cosmos-Hub, an online analysis platform that converts your 16S V3–V4 FASTQ files into interpretable results using a standardized, version-controlled workflow. You receive a QC-filtered ASV table with taxonomy, clear quality summaries, and publication-ready visualizations. Comparative analysis tools are built in, including tables, heatmaps, bar and stacked plots, multiple alpha and beta diversity metrics, abundance distribution views, differential abundance testing, and statistics between groups. Results export as tab-separated tables and PDFs, and each chart can be downloaded as PNG or SVG for downstream use . Learn more about Cosmos-Hub.


Revvity 16S rRNA analysis - online demonstration
Now you can schedule a one-on-one 30-minute online demonstration that will cover everything you need to know about the Cosmos-Hub® which is now available with Revvity’s NEXTFLEX™ 16S rRNA Amplicon-seq Kits. You will be able to see the analysis demonstrated and our experts will be able to answer any questions that you may have.
Automation-ready
Automation protocols are available for the Sciclone™ NGS and NGSx workstations, and the Sciclone NGSx iQ™ workstation supporting walkaway NGS library preparation and higher throughput in 96-well plates while reducing manual steps and variability.
Region options to fit your study
Revvity offers 16S library prep kits for V3–V4, V4, and V1–V3, so you can match the target region to your goals: choose V3–V4 when you want more phylogenetic signal across two adjacent hypervariable domains, pick V4 for the shortest and most widely standardized amplicon that fits common read lengths, or select V1–V3 when your assay or consortium prefers that region for higher precision in certain clades or to align with established SOPs.
Specifications
| Analysis |
Included
|
|---|---|
| Automation Compatible |
Yes
|
| Barcodes |
1 - 48
|
| Product Group |
16S amplicon-seq
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
96 rxns
|
FAQs
-
What sequencing platforms and read lengths are supported?
-
What DNA input range does the kit support?
-
Do I need to buy separate cleanup beads?
-
Which indexes are included, and can I scale higher multiplexing?
-
Which primers are used to target the V3–V4 region?
-
Can I use this kit for archaeal profiling?
-
Why are paired-end 2×300 reads important for this V3–V4 kit?
-
What do the “S” variants include for data analysis?
-
Is the workflow compatible with automation?
Resources
Are you looking for resources, click on the resource type to explore further.
The success of sensitive downstream genomic applications such as qPCR depend on robust and reliable upstream sample preparation...


How can we help you?
We are here to answer your questions.






























