
Gel-free library prep using miRNA isolated from exosomes
Extracellular vesicles are membrane-bound compartments which are being increasingly recognized as important players in cell-to-cell communication. They are secreted by most cell types and are present in many and perhaps all body fluids, including plasma. Extracellular vesicles are classified in different types according to their size.
Key takeaways:
- Gel-free workflow from RNA isolated from exosomes
- Exceptional miRNA discovery
To view the full content please answer a few questions
Introduction
The most characterized are exosomes, which are 50-200 nm in size. Exosomes transport a cargo of different proteins and RNA (exoRNA), mainly mRNA and miRNA. miRNAs in particular are being studied as potential biomarkers for diseases. The fact that they are present in exosomes make them attractive candidates as biomarker molecule as this means that can be collected easily and in a minimally invasive way1-3.
There is a need of a convenient method to construct libraries from exoRNA. The amount of RNA obtained from exosomes is low, with just a few picograms corresponding to miRNA. Based on quantitative analysis this can be as little as 1 miRNA molecule/100 exosomes4, making this a very challenging sample type.
Here, we describe a simplified, commercially available protocol encompassing exoRNA extraction and preparation of exosomal miRNA-seq libraries from serum and bone marrow.
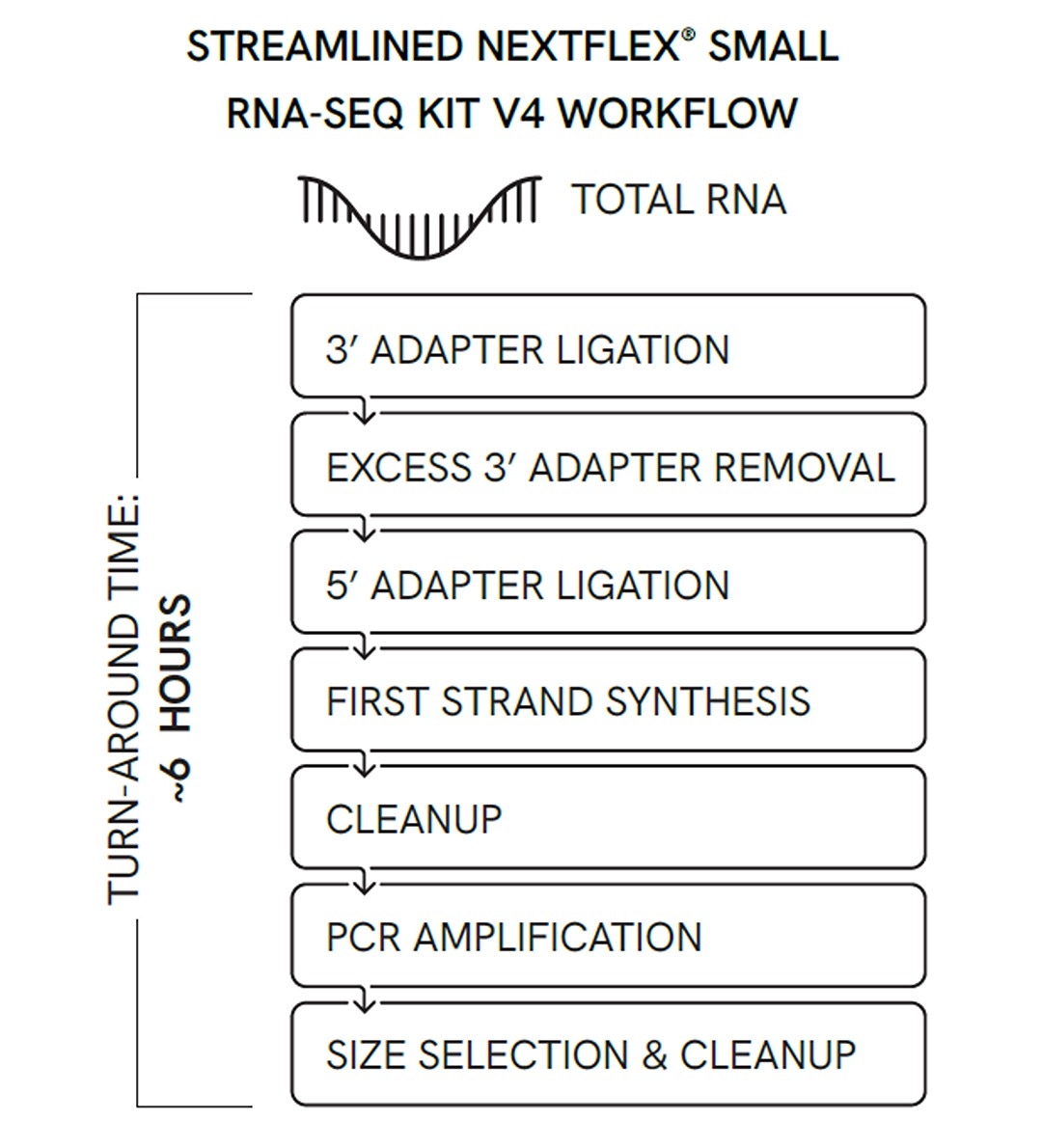
The NEXTFLEX® Small RNA-Seq Kit v4
 NEXTFLEX Small RNA Sequencing Kit V4
is a gel-free solution that was shown previously to produce high rates of miRNA mapping and discovery when working with low input samples such as plasma and serum. Here we wanted to verify that NEXTFLEX® Small RNA-Seq Kit can also be used to investigate microRNAs in a population of purified exosomes. Additionally, the protocol is completely automated protocol from library prep to normalization pooling on the Sciclone® G3 NGS/NGSx workstations
NEXTFLEX Small RNA Sequencing Kit V4
is a gel-free solution that was shown previously to produce high rates of miRNA mapping and discovery when working with low input samples such as plasma and serum. Here we wanted to verify that NEXTFLEX® Small RNA-Seq Kit can also be used to investigate microRNAs in a population of purified exosomes. Additionally, the protocol is completely automated protocol from library prep to normalization pooling on the Sciclone® G3 NGS/NGSx workstations
 Sciclone G3 NGSx iQ Workstation
and Zephyr® G3 NGS workstations
Sciclone G3 NGSx iQ Workstation
and Zephyr® G3 NGS workstations
 Zephyr G3 NGS Workstation
.
Zephyr G3 NGS Workstation
.

Methods
Exosomes from pooled human serum healthy donors and human bone marrow were obtained from System Biosciences. From each sample, 10 µg of purified exosomes was extracted using the NextPrep™ Magnazol™ cfRNA Isolation Kit from Revvity. This corresponds to approximately 2x 109 vesicles5.
12 µL of purified RNA were split into triplicate for sample preparation, equivalent to roughly to 10 pg of input of microRNA per library if we assume 1 miRNA/100 exosomes.
Small RNA libraries were prepared manually using the NEXTFLEX® Small RNA-Seq Kit v4 according to the manufacturer’s instructions except the adapters were used at ¼ dilution. Once libraries were prepared, they were quantified with Thermo Fisher® Scientific Qubit® fluorometer, pooled, and run on an Illumina® MiSeq® platform at 1x75 bp read lengths. Small RNA analysis was performed using a Revvity custom script. Alignment reference was mature miRNA from mirBase v22.1.
Results
The NEXTFLEX® Small RNA-Seq Kit v4 was able to generate gel-free libraries for all samples. After sequencing, filtering, and mapping the data, the proportion of reads that aligned with adapter dimer, tRNA, YRNA, rRNA and miRNA for both sample types were determined. Reads mapping to adapter dimer were below 3% in all cases

Figure 1: Average number of unique microRNA discovered from each sample
Unique miRNA species were identified and quantified in each sample at different thresholds (Figure 1). Even with the relatively low sequencing depth used in this experiment the diversity of the miRNA found in the human serum samples are in agreement with the values reported in the literature6.

Figure 2: Average top 10 miRNA observed with exosomes from bone marrow and pooled serum samples.
Finally, we looked at the top 10 miRNA expressed on each of the replicates of the exosome samples, to illustrate the differences in the content of each type of exosome (Figure 2).
Conclusion
In the present study, we presented a simplified, commercially available protocol encompassing exoRNA extraction and preparation of exosomal miRNA-Seq libraries from serum and bone marrow. This convenient gel-free workflow enables the construction of libraries from exoRNA. Using this workflow, researchers can achieve a high number of reads aligning to mature miRNA and low adapter dimers, even with the very low miRNA inputs characteristic of exosome samples.
References
- Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int J Nanomedicine. 2020 Sep 22;15:6917-6934.
doi: 10.2147/IJN.S264498. PMID: 33061359;
PMCID: PMC7519827. - Yang Y, Huang H, Li Y. Roles of exosomes and exosomederived miRNAs in pulmonary fibrosis. Front Pharmacol. 2022 Aug 11;13:928933. doi: 10.3389/fphar.2022.928933.
PMID: 36034858; PMCID: PMC9403513. - Chuanyun Li, Tong Zhou, Jing Chen, Rong Li, Huan Chen, Shumin Luo, Dexi Chen, Cao Cai, Weihua Li, The role of Exosomal miRNAs in cancer, Journal of Translational Medicine, 20, 1, (2022). https://doi.org/10.1186/s12967- 021-03215-4.
- Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14888-93. doi: 10.1073/pnas.1408301111.
Epub 2014 Sep 29. PMID: 25267620;
PMCID: PMC4205618. - xosome Research | System Biosciences [www.systembio.com/products/exosome-research]
- Shi H, Jiang X, Xu C, Cheng Q. MicroRNAs in Serum Exosomes as Circulating Biomarkers for Postmenopausal Osteoporosis. Front Endocrinol (Lausanne). 2022 Mar 10;13:819056. doi: 10.3389/fendo.2022.819056.
PMID: 35360081; PMCID: PMC8960856.
Featured products

NEXTFLEX Small RNA Sequencing Kit V4
NEXTFLEX small RNA sequencing kit v4は、特許申請中の技術を活用し、IlluminaおよびElementシーケンシングプラットフォーム向けに完全にゲルフリーなsmall RNAライブラリー作成ソリューションを提供します。
- 精製されたmiRNAまたはわずか1ngのトータルRNAから、完全にゲルを使用しないプロトコル。PAGEクリーンアップは不要。
- Sciclone™ G3 NGSx、Sciclone G3 NGSx iQ™、およびZephyr™ G3 NGSワークステーションで自動化対応、完全自動化によるスケーラビリティを実現
- 低バイアスのライゲーションとダイマー形成の抑制によって、処理の難しい生体液(血漿、血清、尿、脳脊髄液)や細胞外小胞からのデータ品質を向上
- 内部ベンチマーキングでは、わずか1 ngのトータルRNAから検出されるユニークなmiRNA種が最大45%増加することが示されており、堅牢な差異発現解析と新規のsmall RNA発見をサポート
- 6時間以内でライブラリーが調製可能、迅速なバイオマーカーおよび機能ゲノミクス研究を支援
- ハイスループットマルチプレキシングおよびIllumina®/Element®プラットフォーム上での最小インデックスホッピングのための384個のユニークデュアルインデックス(UDI)バーコードを含みます。
- 各ロットの厳格なテストを通じて品質を保証
機能研究の一環としてmiRNAレベルを促進または抑制する場合は、Dharmacon™ miRNA modulation試薬をご覧ください。

Sciclone G3 NGSx ワークステーション UVライト搭載
Sciclone™ G3 ワークステーションは、多くの液体分注アプリケーションを自動化するように構成可能です。オープンデッキ設計により、96チャンネルまたは384チャンネルのピペッティングヘッド、オプションの8チャンネルピペッター、さらにはバルク試薬ディスペンサーを組み込むことができます。Revvityの「PING!」非接触液面検出技術により、真空または圧力ろ過を使用してすべてのサンプルが処理されることを保証します。磁気ビーズを使用した分離やデッキ上での多様な振とう、加熱、冷却オプションも、この液体分注機でサポートされています。
特徴および利点
- オープンでモジュラー式の設計ピペッティングヘッド
- 96及び384ウェルプレート密度
- 100 nLから200 μLまでの容量
- 独立した8チャンネルピペッティング機能
- Scicloneグリッパー、バルク試薬モジュール、オンデッキ型マイクロプレートシェーカー、温度制御付きロケーター、陽圧ろ過システムなど、多様なオプションと互換性があります。
- 数百もの異なる構成が可能
自動分注機ではRevvityブランドのピペットチップの使用を推奨します。当社のチップは厳しい品質基準で設計・製造されており、最大限の正確性と精度を保証するとともに、Revvityリキッドハンドラーでの使用を保証された唯一のチップです。
Z-8ピペッティングモジュールをScicloneリキッドハンドラーに追加して、チェリーピッキング、リフォーマット、ノーマライゼーションプロトコルなどを実現します。これにより、独立したZ軸(上下)移動制御を持つ8チャンネルが提供され、異なる容量での吸引と分注が同時に可能です。

Zephyr G3 NGS ワークステーション
Zephyr G3 NGS自動液体分注機プラットフォームは、完全なライブラリー調製自動化や、PCR前およびPCR後のプロセス分離用に特化した、コンパクトで使いやすいシステムです。1日に最大48〜96サンプルを処理可能です。
このワークステーションには以下が含まれます:
- 高精度の96チャンネルピペッティングヘッド
- 統合オフセットグリッパーにより、消耗品デッキの再配置やピペッティング時のフタ保持が可能(汚染の可能性を最小化)
- 部分的なチップロードが可能で、チップの無駄を削減
- オンデッキ温度制御オプション
- オンデッキ振とうオプション
- 廃棄チップシュート
- Maestro™ コントロールソフトウェアとアプリケーションアシスタントユーザーインターフェース
このワークステーションは、DNA、RNA、エクソーム、またはメタゲノミクスのNGSライブラリー調製を行うことができ、40種類以上の検証済みライブラリー調製方法が含まれています。
リキッドハンドラーではRevvityブランドのピペットチップの使用を推奨します。弊社のチップは厳しい品質基準で設計・製造されており、最大限の精度と正確性を保証するとともに、Revvityリキッドハンドラーでの使用を保証された唯一のチップです。
Featured categories

miRNA Analysis
The purification of small RNA species such as microRNA (miRNA) is challenging because of the low concentrations, small fragment sizes (19-22 nucleotides) and tendency to be found in protein complexes.
Exosome/cfRNA Analysis
Extracellular vesicles are membrane-bound compartments which are being increasingly recognized as important players in cell-to-cell communication.

miRNA-seq Analysis
There are different species of small non-coding RNAs (sncRNAs) that play critical roles in various regulatory processes such as transcription, post-transcription, and translation.




























