Explore our solutions

Customer experiences
自動DNA抽出法は、遺伝学研究に革命をもたらしています。このことは、2つの洞察に満ちたウェビナーで示されています。パリのKarine Auribault氏とカンザスのEmily Farrow氏が、病院環境におけるハイスループットDNA抽出技術の導入経験を共有します。これらのプレゼンテーションでは、MLPAやサンガーシーケンシングから、最先端のロングリードシーケンシングに至るまで、幅広い遺伝子検査およびアッセイを可能にするために、効率的な高分子量(HMW)DNA抽出がいかに重要であるかが強調されます。
自動DNA抽出法は、遺伝学研究に革命をもたらしています。このことは、2つの洞察に満ちたウェビナーで示されています。パリのKarine Auribault氏とカンザスのEmily Farrow氏が、病院環境におけるハイスループットDNA抽出技術の導入経験を共有します。これらのプレゼンテーションでは、MLPAやサンガーシーケンシングから、最先端のロングリードシーケンシングに至るまで、幅広い遺伝子検査およびアッセイを可能にするために、効率的な高分子量(HMW)DNA抽出がいかに重要であるかが強調されます。
Suggested links:

Webinar review: High quality DNA for genetic testing

Unleashing the power of genomics: A webinar review
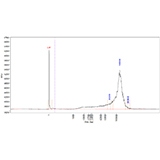
長鎖シーケンシング用の高分子量DNA抽出
アプリケーションノートでは、効率的な高分子量(HMW)DNA抽出について詳しくご紹介しています。DNA収量、純度、完全性に基づいて、2つの自動核酸抽出システムによる血液からの抽出性能を比較しました。
アプリケーションノートでは、効率的な高分子量(HMW)DNA抽出について詳しくご紹介しています。DNA収量、純度、完全性に基づいて、2つの自動核酸抽出システムによる血液からの抽出性能を比較しました。
Suggested links:
Comparison of automated nucleic acid purification systems HMW DNA extraction efficiency

バイオバンキング
血液、バフィーコート、スワブ、唾液など、さまざまなサンプルタイプにおけるハイスループット抽出の豊富な専門知識を活かし、私たちはバイオバンキング分野でグローバルな存在感を確立しています。chemagicによる核酸分離は、アーカイブ品質の検体を保証し、一次サンプルの取り扱いから下流の品質管理およびアッセイ設定まで、ワークフロー全体の自動化を可能にすることで、さまざまな集団遺伝学コホート研究において信頼されるパートナーとしての地位を築いています。
血液、バフィーコート、スワブ、唾液など、さまざまなサンプルタイプにおけるハイスループット抽出の豊富な専門知識を活かし、私たちはバイオバンキング分野でグローバルな存在感を確立しています。chemagicによる核酸分離は、アーカイブ品質の検体を保証し、一次サンプルの取り扱いから下流の品質管理およびアッセイ設定まで、ワークフロー全体の自動化を可能にすることで、さまざまな集団遺伝学コホート研究において信頼されるパートナーとしての地位を築いています。
O私たちの貢献は、Mayo Clinicの「All of Us Research Programme」7、German National Cohort/NAKO Gesundheitsstudie8、および英国の100,000 Genomes Projectを含むイニシアチブなど、注目すべきプロジェクトにまで及びます。
Suggested links:
Dried blood spots for advanced DNA sequencing
Automated nucleic acid isolation fulfilling biobanking needs

High quality DNA isolation suitable for ultra rapid sequencing

オックスフォード・ナノポアを使用した全ゲノムシーケンシング
ある先進的なラボが、chemagicで精製された高分子量DNA抽出を取り入れることで、Oxford Nanoporeのライゲーションベース全ゲノムシーケンシングワークフローをどのように強化したかをご覧ください。
この最適化は、前処理段階のサンプル調製が、後工程のシーケンシング品質およびデータの完全性に与える重要な影響を示しています。
ある先進的なラボが、chemagicで精製された高分子量DNA抽出を取り入れることで、Oxford Nanoporeのライゲーションベース全ゲノムシーケンシングワークフローをどのように強化したかをご覧ください。
この最適化は、前処理段階のサンプル調製が、後工程のシーケンシング品質およびデータの完全性に与える重要な影響を示しています。
References:
- Lang K, Wagner I, Schöne B, et al. ABO allele-level frequency estimation based on population-scale genotyping by next generation sequencing. BMC Genomics. 2016;17:374. Published 2016 May 20.
- Beyter D, Ingimundardottir H, Oddsson A, et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nature Genetics. 2021 Jun;53(6):779-786.
- Steiert TA, Fuß J, Juzenas S, et al. High-throughput method for the hybridisation-based targeted enrichment of long genomic fragments for PacBio third-generation sequencing. NAR Genom Bioinform. 2022;4(3):lqac051. Published 2022 Jul 13.
- Schmidt J, Berghaus S, Blessing F, et al. Genotyping of familial Mediterranean fever gene (MEFV)-Single nucleotide polymorphism-Comparison of Nanopore with conventional Sanger sequencing. PLoS One. 2022;17(3):e0265622. Published 2022 Mar 17.
- Watson CM, Crinnion LA, Hewitt S, et al. Cas9-based enrichment and single-molecule sequencing for precise characterization of genomic duplications. Lab Invest. 2020;100(1):135-146.
- Watson CM, Crinnion LA, Simmonds J, Camm N, Adlard J, Bonthron DT. Long-read nanopore sequencing enables accurate confirmation of a recurrent PMS2 insertion-deletion variant located in a region of complex genomic architecture. Cancer Genet. 2021;256-257:122-126.
- Valentin N, Camilleri M, Carlson P, et al. Potential mechanisms of effects of serum-derived bovine immunoglobulin/protein isolate therapy in patients with diarrhea-predominant irritable bowel syndrome. Physiol Rep. 2017;5(5):e13170.
- Kousathanas A, Pairo-Castineira E, Rawlik K, et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. 2022;607(7917):97-103.