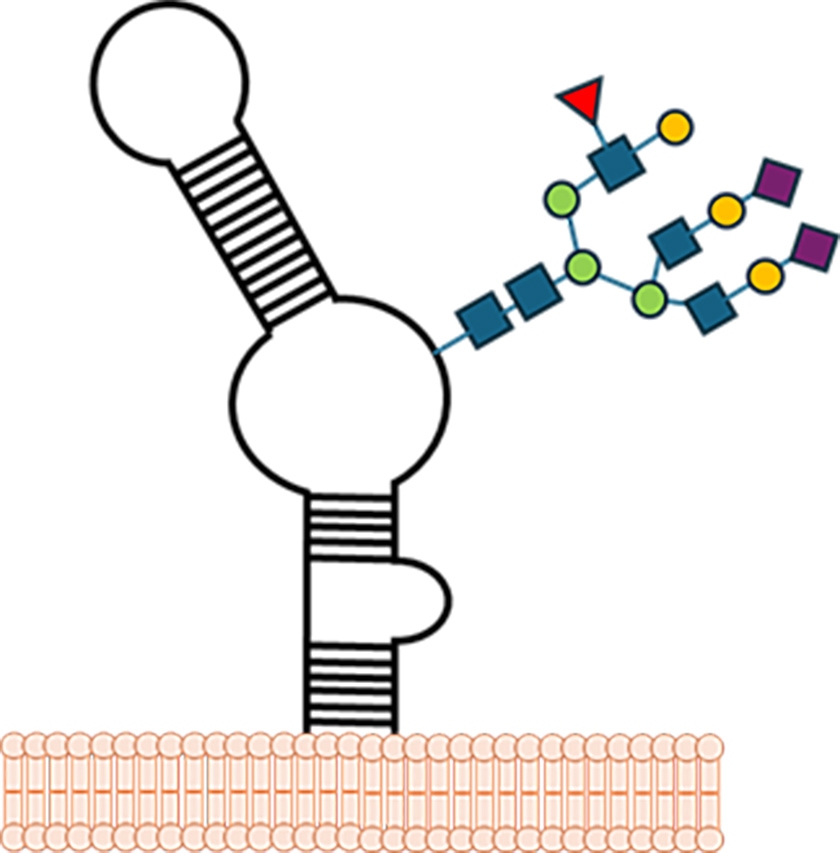
For decades, biologists believed that glycosylation, the covalent addition of complex sugars (glycans), was exclusive to proteins and lipids. This changed in 2021, when Flynn et al. published a landmark study showing that small RNAs can also be glycosylated. These molecules, termed glycoRNAs, were observed across multiple mammalian cell types and species1.
GlycoRNAs were found to carry sialylated N-glycans, sugar chains terminated with sialic acid residues. Sialylation refers to the addition of sialic acid, a negatively charged monosaccharide, to the ends of glycan chains. This modification is critical in cell–cell communication and immune recognition: sialylated glycans often act as molecular “flags” read by receptors such as sialic acid-binding immunoglobulin-like lectins (Siglecs), which tune immune responses and prevent autoimmunity2. Finding sialylation on RNA fundamentally challenged our understanding of both glycobiology and RNA biology (Figure 1).

Figure 1. Scheme of a glycoRNA on the surface of a cell. Modified from Disney (2021)3.
Given the extraordinary nature of this discovery, the initial reports sparked debate. Were these glycoRNAs real biological entities, or artifacts of preparation? A follow up study by Flynn's lab addressed this by using advanced chemical labelling tools, northern blots, and mass spectrometry, further supporting glycoRNAs as authentic biological molecules4.
Growing evidence suggests that glycoRNA are detectable both in vitro and in vivo, particularly on the cell surface. Among small RNAs, Y RNAs often represent the largest fraction of glycoRNA species, followed by tRNAs, snRNAs, snoRNAs, and others1.
Advanced mass spectrometry-based workflows have profiled N-glycans on RNA quantitatively across tissues. These analyses revealed that glycoRNA glycans differ significantly in composition from protein-bound glycans and exhibit tissue-specific abundance patterns, suggesting that glycoRNA levels may vary based on cellular context and physiological state5.
Finally, in a transcriptome-wide draft of human glycoRNA glycan profiles, the number of distinct glycan species was limited, yet they were present at relatively high abundance levels. This suggests that although the glycan variety is narrow, their representation on RNA in the glycoRNA pool is significant6.
Mechanism of RNA glycosylation
A pivotal study by Xie et al identified the modified nucleotide acp³U (3-(3-amino-3-carboxypropyl)uridine) as the key site for N-glycan attachment on tRNAs and related RNA. It is worth noting that in tRNA this modification is commonly present in eukaryotes and bacteria5.
The authors proposed a 3-step model pathway revealing how classical protein-focused glycosylation machinery also extends its reach to RNA substrates. According to this model first acp³U is introduced during tRNA maturation in the nucleus/cytosol. Then the tRNA enters the secretory pathway, allowing the N-glycosylation machinery to attach syalyated glycans. Finally, the resulting glycoRNAs are displayed on the cell surface4.
Functional implications
GlycoRNAs appear to extend RNA's functional repertoire into extracellular and immunological realms, acting as novel mediators in cell–cell communication and immune regulation
- Immune modulation via Siglecs: GlycoRNAs, decorated with sialylated N‑glycans, are recognized by Siglec receptors, immune regulatory proteins that bind sialic acid–terminated glycans8. This interaction may tune immune activation, tolerance, and inflammatory responses, positioning glycoRNAs as RNA-based immune modulators.
- Surface signaling and cellular entry: GlycoRNAs have been found to assemble into specialized membrane domains, interacting with RNA-binding proteins and facilitating entry of cell-penetrating peptides, a potential new route for molecular signaling or therapeutic delivery9.
- Extracellular vesicle-associated biomarkers: GlycoRNAs carried in small extracellular vesicles can be profiled using drFRET, yielding remarkable diagnostic performance: 100% accuracy distinguishing cancer versus control, and ~90% accuracy in subclassifying cancer types within patient cohorts10.
These insights support the idea that glycoRNAs constitute a relevant, regulated layer of extracellular RNA biology, with intriguing links to immune signaling, disease states, and diagnostic potential.
Detecting and characterizing GlycoRNAs
Small RNA sequencing
Because glycoRNAs are derived from small RNAs, sequencing these fractions has been essential to identify their RNA backbones. When combined with glycan-specific enrichment (lectins, click chemistry, or rPAL), small RNA sequencing
 NEXTFLEX Small RNA Sequencing Kit V4
reveals exactly which RNAs are glycosylated. In fact Flynn et al. confirmed in their landmark paper that glycoRNAs correspond to intact small RNAs, not degradation products, using sequencing. Pairing NGS with chemical labeling and/or mass spectrometry seems to be key to achieving comprehensive glycoRNA maps in the near future.
NEXTFLEX Small RNA Sequencing Kit V4
reveals exactly which RNAs are glycosylated. In fact Flynn et al. confirmed in their landmark paper that glycoRNAs correspond to intact small RNAs, not degradation products, using sequencing. Pairing NGS with chemical labeling and/or mass spectrometry seems to be key to achieving comprehensive glycoRNA maps in the near future.
Metabolic labeling + northwestern blotting
Cells can be fed unnatural sugars (e.g., Ac₄ManNAz), which become incorporated into glycans. These modified glycans can then be “clicked” to detection reagents. Coupled with northwestern blots, this method first revealed cell-surface glycoRNAs. A detailed step-by-step protocol has been recently published11.
rPAL (RNA-optimized periodate oxidation and aldehyde ligation)
The rPAL technique exploits periodate to oxidize vicinal diols within RNA, creating aldehydes that can be ligated to tagging molecules. Compared to metabolic labeling, rPAL provides a ~25-fold increase in sensitivity and better signal recovery12.
Lectin-based proximity labeling
Lectins are proteins that bind specifically and reversibly to carbohydrate structures. Lectins such as Wheat Germ Agglutining (binding sialic acid/GlcNAc) enrich glycoRNAs for downstream analysis12.
drFRET for extracellular vesicle GlycoRNA profiling
The drFRET assay uses dual probes recognizing both RNA and glycan moieties, enabling ultrasensitive detection of glycoRNAs in biofluids from as little as 10 µL of biofluid. In clinical studies, drFRET was combined with Small RNA sequencing to correlate glycoRNA presence with specific small RNA species, supporting its potential for diagnostic stratification10.
Conclusions and outlook
GlycoRNAs are small RNAs covalently coated with glycans, prominently sialylated N-glycans. Other glycans may be present. In fact, a recent study suggests that O-glycosylation also contributes to glycoRNA biogenesis in some systems12.
Their discovery reshapes RNA biology and introduces new paradigms for cell communication and immune regulation. For scientists and innovators in RNA research, glycoRNAs represent a frontier comparable to the discovery of microRNAs or RNA modifications two decades ago. As detection tools evolve, we may uncover glycoRNAs as key regulators in health, disease, and therapeutic development.
References:
- Flynn, R.A. et al (2021). Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell, 184(12), 3109-3024. doi: 10.1016/j.cell.2021.04.023.
- Zhu, W., et al. (2024). Biological function of sialic acid and sialylation in human health and disease. Cell Death Discov. 10, 415. Doi: 10.1038/s41420-024-02180-3.
- Disney, M.D. (2021). A glimpse at the glycoRNA world. Cell, 184(12), 3080 – 3081. doi: 10.1016/j.cell.2021.05.025.
- Xie, Y. et al (2024). The modified RNA base acp3U is an attachment site for N-glycans in glycoRNA Cell, 187(19), 5228-5237. DOI: 10.1016/j.cell.2024.07.044.
- Xie, Y., et al. (2025). Development and application of GlycanDIA workflow for glycomic analysis. Nat Commun 16, 7075. Doi: 10.1038/s41467-025-61473-y.
- Bi, M., et al. (2023). A draft of human N-glycans of glycoRNA. bioRxiv [preprint] doi: 10.1101/2023.09.18.558371
- Takakura, M. et al. (2019). Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat Commun 10, 5542. Doi:10.1038/s41467-019-13525-3.
- Clyde, D. Sugar-coated RNAs (2021). Nat Rev Genet 22, 480. Doi: 10.1038/s41576-021-00388-y.
- Perr, J. et al. (2025). RNA-binding proteins and glycoRNAs form domains on the cell surface for cell-penetrating peptide entry. Cell, 188(7), 1878-1895. doi: 10.1016/j.cell.2025.01.040
- Ren, T., et al (2025). FRET imaging of glycoRNA on small extracellular vesicles enabling sensitive cancer diagnostics. Nat Commun. 16(1):3391. doi: 10.1038/s41467-025-58490-2.
- Li, L. et al (2024) Protocol for detecting glycoRNAs using metabolic labeling and northwestern blot. STAR Protoc. 5(4):103321. doi: 10.1016/j.xpro.2024.103321.
- Porat, J., et al (2024). O-glycosylation contributes to mammalian glycoRNA biogenesis. bioRxiv [Preprint]. doi: 10.1101/2024.08.28.610074


































