
NEXTFLEX UDI Barcodes (10NT, 769-1152)



| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | バーコード |



Product information
Overview
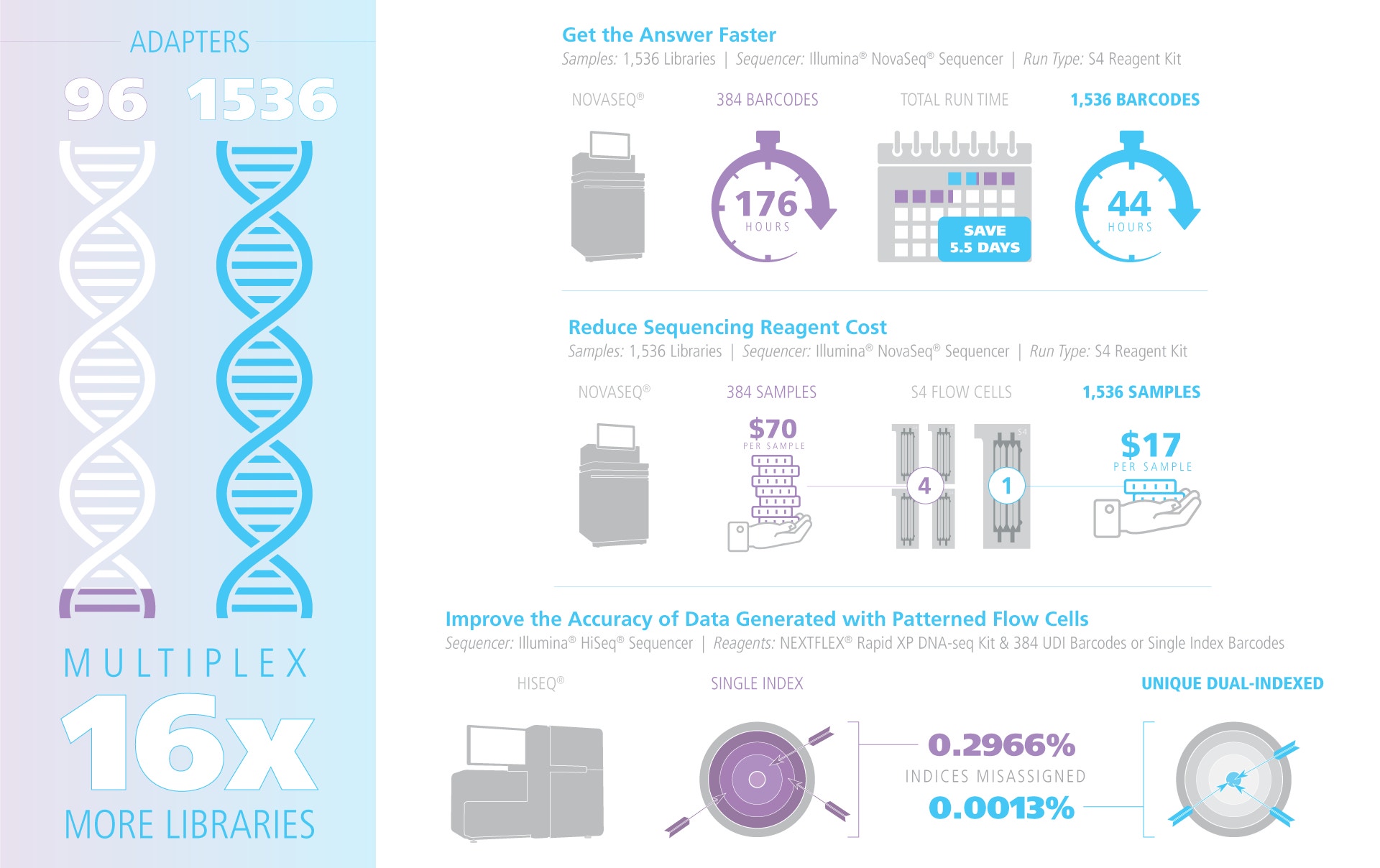
- Up to 1,536 unique i5 / i7 pairs (10NT) ready to ship
- Compatible with PCR-free and PCR-amplified workflows
- ≥ 3-base Hamming distance across the entire set for robust error-correction and balanced base diversity on 2- & 4-color chemistries
- Designed to minimize index hopping that can be present with patterned flow cells.
- Compatible with Revvity library prep and TruSeq style, ligation-based workflows
- Every lot is functionally validated and tested for index purity by sequencing in an ISO 9001 facility
- Optional NEXTFLEX Universal Blockers boost on-target hybrid-capture efficiency when used with these UDIs.
Additional product information
NGS Multiplexing Performance
Scale from 2 to 1,536 libraries in a single NovaSeq® (Illumina) or AVITI® (Element)flow cell. The 10NT UDI set supports barcode rotation between runs, eliminating carry-over and virtually stopping index hopping on patterned flow cells. Adapters work out-of-the-box with all TruSeq-style library-prep kits, including NEXTFLEX, Illumina, Watchmaker, Twist library prep kit s, whether PCR-free or PCR-amplified.
“Revvity has a set of 1536 validated UDI adapters that we have been regularly using for 5 years. Using the Illumina® NovaSeq® X Plus with this UDI set from Revvity we can sequencing 12,288 samples across 8 lanes of the 10B or 25B flow cells. This dramatically lowers our sequencing cost per sample!”
-Dr. Charlie Johnson, Director of Texas A&M Agri Life Genomics and Bioinformatics Service
NGS Adapters with High Data Integrity
Each NEXTFLEX UDI adapter has a dedicated 10NT i7 & i5 sequence separated from every other pair by a ≥ 3-base Hamming distance, enabling single-base error correction during demultiplexing. Because the adapter already contains the full flow-cell and primer-binding sites, PCR enrichment is optional. On patterned flow cells (Illumina NovaSeq®, HiSeq® 3000/4000, HiSeq® X, Element AVITI™) the use of true, unique dual indexes cuts index-hopping artefacts to background levels by removing mis-assigned reads. Every lot is sequenced for ≥ 99.9 % index purity and functional ligation efficiency, ensuring data integrity run after run.

Specifications
| Automation Compatible |
Yes
|
|---|---|
| Barcodes |
769 - 1152
|
| Format |
2 rxns per well
|
| Product Group |
Barcodes
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
768 rxns
|
References
Cellular & organismal transcriptomics / functional genomics
- Estermann, M. (2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. preLights. https://doi.org/10.1242/prelights.18906
- Veenvliet, J. V., Bolondi, A., Kretzmer, H., Haut, L., Scholze-Wittler, M., Schifferl, D., … Herrmann, B. G. (2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science, 370(6522), eaba4937. https://doi.org/10.1126/science.aba4937
- Hirose, K., Chang, S., Yu, H., Wang, J., Barca, E., Chen, X., … Huang, G. N. (2019). Loss of a novel striated muscle-enriched mitochondrial protein Coq10a enhances postnatal cardiac hypertrophic growth. bioRxiv. https://doi.org/10.1101/755793
- León, K. E., Buj, R., Lesko, E., Dahl, E. S., Chen, C.-W., Imamura, Y., … Aird, K. M. (2020). DOT1L modulates the senescence-associated secretory phenotype through epigenetic regulation of IL1A. bioRxiv. https://doi.org/10.1101/2020.08.21.258020
- Dubey, S. K., Dubey, R., Jung, K., Hernandez, A. G., & Kleinman, M. E. (2025). Deciphering age-related transcriptomic changes in the mouse retinal pigment epithelium. Aging (Albany NY), 17(3), 657–684. https://doi.org/10.18632/aging.206219
- Pujari, A. N., & Cullen, P. J. (2024). Modulators of MAPK pathway activity during filamentous growth in Saccharomyces cerevisiae. G3 (Bethesda), 14(6), jkae072. https://doi.org/10.1093/g3journal/jkae072
- Miura, H., Takahashi, S., & Shibata, T. (2020). Mapping replication timing domains genome wide in single mammalian cells with single-cell DNA replication sequencing. Nature Protocols, 15(12), 4058–4100. https://doi.org/10.1038/s41596-020-0378-5
- Gardner, E. J., Prigmore, E., Gallone, D., Danecek, P., Samocha, K. E., Handsaker, J., … Hurles, M. E. (2019). Contribution of retrotransposition to developmental disorders. Nature Communications, 10(1), 4630. https://doi.org/10.1038/s41467-019-12520-y
Microbial, environmental, and pathogen genomics / metagenomics
- Adolfi, A., Gantz, V. M., Jasinskiene, N., Lee, H.-F., Hwang, K., Terradas, G., … James, A. A. (2020). Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nature Communications, 11, 5553. https://doi.org/10.1038/s41467-020-19426-0
- Gaeta, N. C., Bean, E., Miles, A. M., de Carvalho, D. U. O. G., Alemán, M. A. R., Carvalho, J. S., … Ganda, E. (2020). A cross-sectional study of dairy cattle metagenomes reveals increased antimicrobial resistance in animals farmed in a heavy metal contaminated environment. Frontiers in Microbiology, 11, 590325. https://doi.org/10.3389/fmicb.2020.590325
- Kant, P., Brohier, N., Mann, R., Rigano, L., Taylor, R., Wong-Bajracharya, J., … Constable, F. (2023). High-quality full genome assembly of historic Xylella fastidiosa strains from ICMP collection using a hybrid sequencing approach. Microbiology Resource Announcements, 12(11), e00536-23. https://doi.org/10.1128/MRA.00536-23
- Arras, S. D. M., Sibaeva, N., Catchpole, R. J., Horinouchi, N., Si, D., Rickerby, A. M., … Poole, A. M. (2023). Characterisation of an Escherichia coli line that completely lacks ribonucleotide reduction yields insights into the evolution of parasitism and endosymbiosis. eLife, 12, e83845. https://doi.org/10.7554/eLife.83845
- Larkin, A. A., Brock, M. L., Fagan, A. J., Moreno, A. R., Gerace, S. D., Lees, L. E., … Martiny, A. C. (2025). Climate-driven succession in marine microbiome biodiversity and biogeochemical function. Nature Communications, 16(1), 3926. https://doi.org/10.1038/s41467-025-59382-1
- Klauer, R. R., Silvestri, R., White, H., Hayes, R. D., Riley, R., Lipzen, A., … Blenner, M. A. (2024). Hydrophobins from Aspergillus mediate fungal interactions with microplastics. bioRxiv. https://doi.org/10.1101/2024.11.05.622132
- Starr, T. N., Greaney, A. J., Hilton, S. K., Crawford, K. H. D., Navarro, M. J., Bowen, J. E., … Bloom, J. D. (2020). Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. bioRxiv. https://doi.org/10.1101/2020.06.17.157982
FAQs
-
Are the UDI barcodes sequence-verified and functionally validated?
-
What is the lead time when I order the NEXTFLEX unique dual index barcodes?
-
Must I perform paired-end sequencing with NEXTFLEX Unique Dual Index Barcodes?
-
Can I use the NEXTFLEX Unique Dual Index Barcodes in single index experiments?
-
What are the storage and shipping conditions?
-
Do the UDIs work with PCR-free workflows?
-
Can I combine just a subset of the indexes in one run?
-
How do unique dual indexes mitigate index hopping?
Resources
Are you looking for resources, click on the resource type to explore further.
This flyer describes the benefits of the NEXTFLEX® 1536 UDI Barcode Adapter Set.


How can we help you?
We are here to answer your questions.






























