Ever wondered how your drugs work in your in vitro model? By employing functional genomic screening strategies, researchers can understand the impact at the gene level of therapeutic candidates, facilitating the progress of a candidate to the clinic.
The genomic exploration toolkit
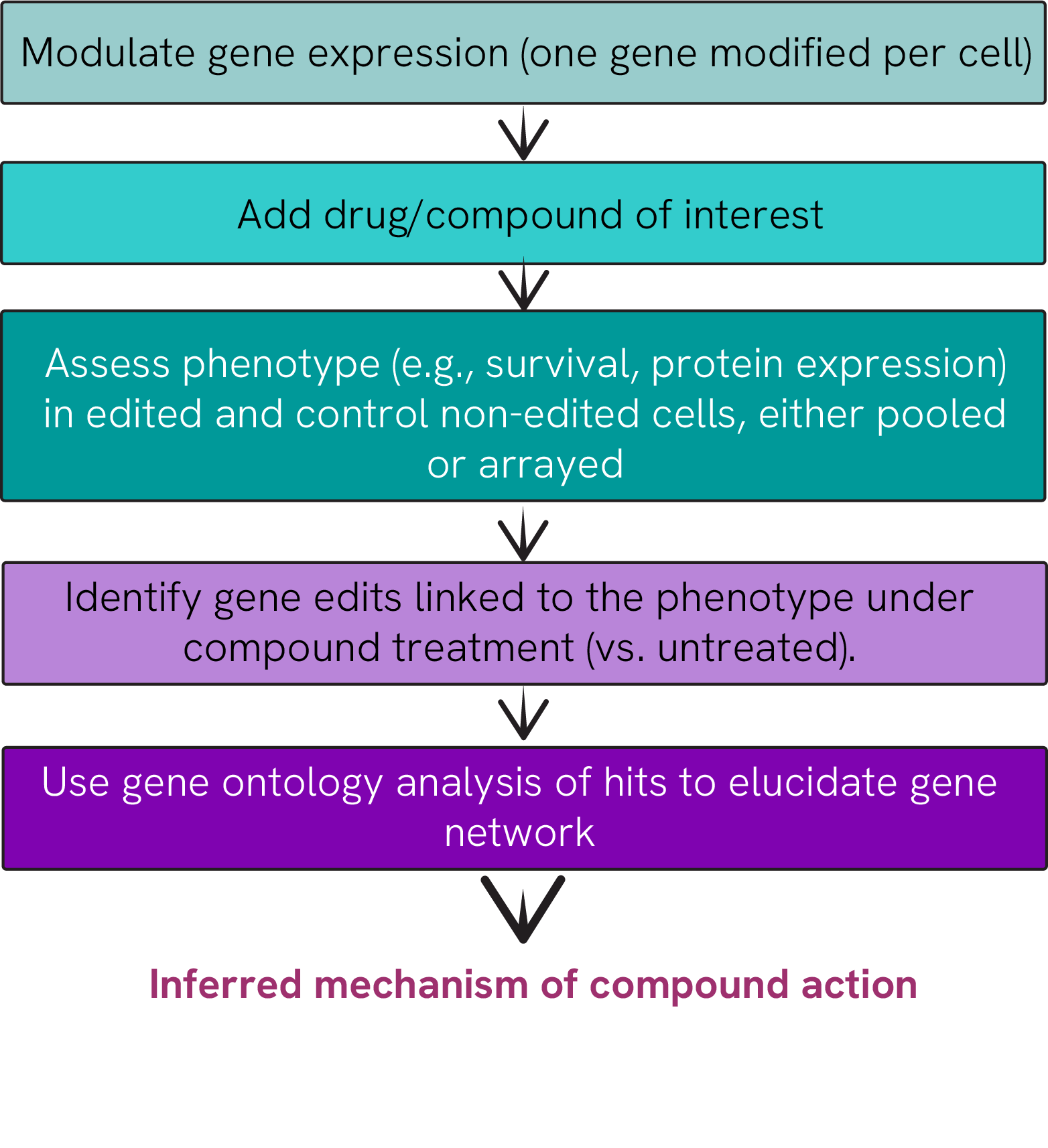
The scientific landscape has evolved significantly from studying individual genes in isolation. With technologies like RNAi and CRISPR, researchers can now analyze thousands of genes simultaneously to observe their functions. This approach, known as Functional Genomic Screening (FGS), provides comprehensive insights into gene function and drug mechanisms of action (MoA). At Revvity, we've developed considerable expertise in providing both tools and services for these sophisticated genetic investigations, enabling researchers to efficiently uncover cellular mechanisms. (Figure 1).

Figure 1. Scientific workflow to discern the mechanism of action.
In this blog we describe the five key factors a researcher should consider in order to design an effective screen.
Five key considerations for effective screening design
- Select the appropriate model system
Many aspects of a screening assay can be tailored to help discern a compound’s mechanism of action (MoA). When investigating a novel compound, a scientist must first decide whether to use a simple 2D cell culture model or a more complex 3D system. A 2D model might involve a monolayer of cells—such as an epithelial cancer cell line derived from the relevant diseased tissue where the compound is believed to act. 3D models incorporate multiple cell layers arranged in a three-dimensional matrix, offering a more physiologically relevant representation of in vivo biological systems.
- Determine the optimcal technology approach
The second key consideration is which FGS technology to utilize. Several options are available:
- CRISPRko for complete gene knockout
- CRISPRi or siRNA for gene expression reduction
- CRISPRa for gene expression enhancement
Many researchers find it helpful to combine these technologies to confirm results in different ways known as an orthogonal approach.
Loss-of-function technologies, such as CRISPR knockout (CRISPRko), CRISPR interference (CRISPRi), and small interfering RNA (siRNA), enable researchers to assess cellular phenotypes after gene ablation or impairment of gene expression. CRISPRko specifically targets and knocks out a gene, while CRISPRi and siRNA lead to reduced gene expression. This capability makes CRISPRi and siRNA more closely aligned with modeling druggability, providing researchers with alternative methods to CRISPRko.
In fact, dual loss-of-function screening—where both CRISPRko and CRISPRi screenings are conducted simultaneously—can enhance confidence in understanding the mechanisms of action. This approach confirms shared targets identified by both methods and allows for the discovery of unique targets associated with each technology, offering a more comprehensive understanding of gene function.
A gain-of-function approach can be achieved using CRISPR activation (CRISPRa), allowing researchers to assess phenotypes in the presence of gene overexpression. Dual-direction screening involves pairing CRISPRa with loss-of-function technologies such as CRISPRko or CRISPRi . By combining parallel studies of gene ablation or gene expression impairment with overexpression, we can investigate opposite phenotypic effects on genes and gain deeper insights into pathway identification and mechanisms of action.
For instance, Revvity has previously utilized all three CRISPR technologies—CRISPRko, CRISPRi, and CRISPRa—in a genome-wide screen to identify genes that contribute to both sensitivity and resistance to the BRAF inhibitor, vemurafenib. This approach facilitated the elucidation of MoA through systematic identification of hits and evaluation of gene networks concerning the drug's function1.
If more complex network analysis is needed, Revvity also offers dual-guide perturbation screens services, which allow for the simultaneous perturbation of two genes of interest in each cell.
- Define the scope of your investigation
When deciding on a genomic analysis approach, consider whether a whole-genome analysis using established off-the-shelf libraries, such as those offered by Revvity, is suitable, or if a targeted approach focusing on specific candidate genes would be more appropriate.
It is also important to consider the number of guides per gene, as this will impact the statistical power and confidence during hit calling and data analysis. For example, in statistical analyses of pooled screens that identify gene modifications leading to drug sensitivity—where affected cells become depleted from the pooled population—a higher number of guides can be advantageous.
- Establish clear phenotypic readouts
The fourth consideration is to determine what you will measure to assess the function of a compound. This could involve evaluating survival and death phenotypes or more specific markers, such as changes in protein expression in response to drug exposure. In other words, decide how you will measure the compound's function and define what the expected functional readout is.
- Choose between arrayed and pooled formats
Screening analysis can be conducted in two main formats: arrayed and pooled. In arrayed screens, individual genes are examined separately in distinct wells of a multiwell plate, with each well treated with a specific gene modulator (such as guide RNA or siRNA). In contrast, pooled screens investigate all candidate genes simultaneously, with each cell in the pool receiving a single modification.
When deciding which format to use, consider your research question and desired outcomes. Arrayed screens provide immediate phenotypic information and can capture multiple phenotypic readouts. They are particularly beneficial for studying non-dividing cells, such as neurons, and in co-culture systems where scientists may want to assess the phenotype of a cell that is not being directly edited.
On the other hand, pooled screens are better suited for simpler readouts, like cell viability. They offer an unbiased approach to whole-genome screening and utilize next-generation sequencing as a readout. The complexity of pooled screen readouts can also be enhanced using technologies such as high content imaging and fluorescence-activated cell sorting (FACS), which enable the investigation of phenotypes through antibody staining or fluorescently tagged proteins (dig deeper in our blog on making the right choice between arrayed or pooled library).
Interpreting your results
The analysis following your screen involves correlating genetic changes with drug function and phenotypic alterations. This process can be carried out through several steps: identifying potential candidates or "hits," analyzing gene networks, validating hits, and conducting hypothesis-testing experiments. For an example of how we might utilize functional genetic screening to explore the mechanism of action of a hypothetical novel therapeutic compound that induces cell death, refer to Figure 2.

Figure 2. CRISPRko screen to investigate a hypothetical novel therapeutic compound. The compound induces cell death in a specific cancer cell type. If we want to discern the MoA of patient resistance to the activity of this compound, we can conduct a whole genome CRISPRko screen which will identify gene deletions resulting in cell survival (rather than the intended cell killing by the compound). Further validation and investigation of these candidates might unveil the mechanism of drug function and indeed, drug resistance. (1) The varying phenotypes following differential genetic perturbation (in this example gene knockout) and drug exposure. (2) Potential mechanism of the drug. (3) Mechanism of action of gene network and drug on phenotype.
Conclusion
CRISPR screening offers valuable insights into understanding the mechanisms of action of drugs or compounds. When conducting these experiments, it is essential to consider several factors based on the type of phenotypic readout and the overall scope and scale of the investigation. Additionally, utilizing a combination of different CRISPR technologies, including those that can induce gene repression and overexpression in parallel experiments, can enhance confidence in identifying significant hits and the genetic networks involved in drug function.
If you are looking for help with a functional genomic screening project, keep reading to see how our preclinical services team can support you.
References:
- le Sage C, Lawo S, Panicker P, et al. Dual direction CRISPR transcriptional regulation screening uncovers gene networks driving drug resistance. Sci Rep. 2017;7(1):17693.


































